This article has been translated into العربية, বাংলা, Català, Dansk, Deutsch, Ελληνικά, Español, فارسی, Suomi, Français, हिन्दी, Magyar, Italiano, 日本語, 한국어, Mакедонски, Bahasa Melayu, Nederlands, Polski, Română, Русский, Slovenščina, Српски, Svenska, தமிழ் and 汉语.
COVID-19 HGI Results for Data Freeze 3 (July 2020)
September 25, 2020
Brooke Wolford and Kumar Veerapen, on behalf of the COVID-19 HGI
Disclaimers: First, please know this research is ongoing. While we are already making discoveries, we need more samples to have a robust understanding of the genetic contribution to COVID-19 outcomes. The more samples we add to our study, the more confident we will be that the patterns we observe exist and are representative across different groups of patients. Second, we are also not able to tell your probability of having severe COVID-19 given your genetics. Users of our results should not use our findings to diagnose COVID-19 patients by their genotype and should always talk to a medical professional to guide medical choices. Finally, if any vocabulary here is unfamiliar, please send us an email at hgi-faq@icda.bio—we’d be happy to update the information here to provide more clarity. In the coming weeks, additional information explaining concepts or terminology will be made available. In the interim, do look at this resource to review the basics of genetics.The COVID-19 pandemic has affected the everyday lives of societies around the world. Scientists around the world are working hard to better understand the virus and disease. We represent one such group—the COVID-19 Host Genetics Initiative (HGI)—an international team of geneticists focused on identifying human genetic variation that influences responses to SARS-CoV-2 infection and its subsequent disease, COVID-19. Working together, we are curious what parts of a person’s DNA can influence whether someone develops COVID-19 and if they do, how sick they get.
The COVID-19 HGI study design
In our study, we are comparing genetic variation between cases, people who are hospitalized and also have a positive test for SARS-CoV-2, with controls who are people from the general population who do not have a positive test for COVID-19. This comparison is called a Genome Wide Association Study, or GWAS. Check out this video or infographic for an illustrated explanation of GWAS! As of July 2020, we have combined results from eight different studies for a total of 3,199 cases and 897,488 controls.
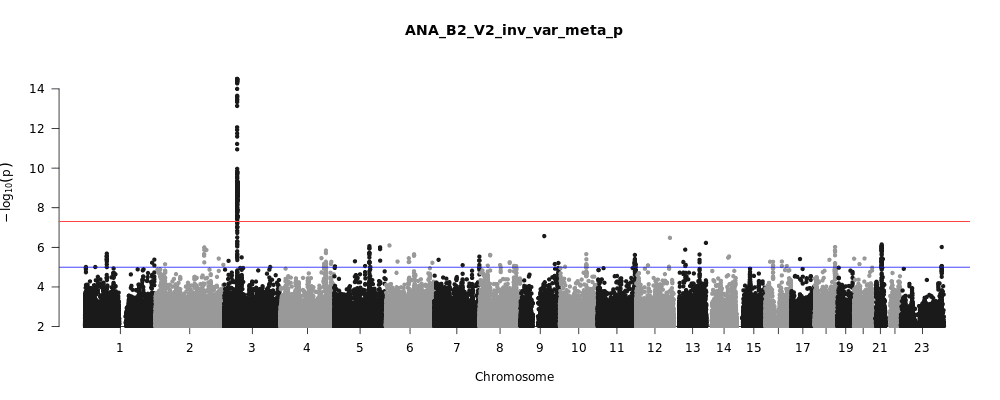
The COVID-19 HGI discovers genetic variation associated with COVID-19 severity
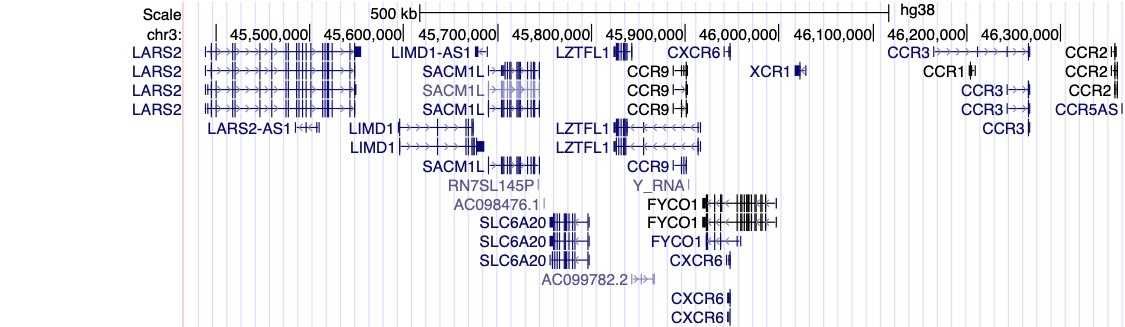
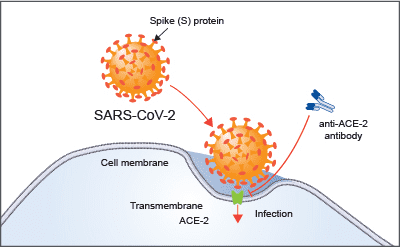
Figure 1, above, shows a visual summary of the most recent results from the COVID-19 HGI. This is called a Manhattan plot, please see the footnote for a full description of this visualization. In short, a Manhattan plot is used to visualize associations between a trait (e.g., COVID-19) and genetic variants across the entire genome. We observe one statistically significant region on chromosome 3 (notice the dotted vertical line above chromosome 3, as indicated on the horizontal x-axis). Sometimes, a region includes multiple genes that are close to each other. It will take additional research to narrow down the statistically significant region to the specific gene involved in COVID-19 severity. The region identified on chromosome 3 overlaps with multiple genes (see all the gene names listed in Figure 2). It is not clear which specific gene within this narrow region is associated with COVID-19 severity. However, we do have some interesting leads! There are several chemokine-related genes in this region, such as CXCR6 and CCR1. Chemokines control the movement of immune cells and are critical for the innate immune system to function properly. The gene SLC6A20 is also in this region, and it makes a protein which is known to bind to ACE2. The ACE2 protein is like a door that the virus SARS-CoV-2 uses for entry into our cells (Figure 3). This means that it’s possible genetic variation in SLC6A20 is influencing viral entry! These results from our discovery of genetic associations are just the first step in the research process.
Comparing our results to those of other studies
You may have heard in the news that blood type appears to be associated with COVID-19, with type A correlating with higher risk and type O being protective. A recent journal article appearing in the New England Journal of Medicine (NEJM) described a genetic association analysis for severe COVID-19 (e.g. hospitalization with respiratory failure) in 1,980 individuals from Italy and Spain (and replicated by 23andMe as well). In this study, the ABO blood group gene on chromosome 9 seems to be significantly associated with COVID-19. However, this study used blood donors as a control group in their analysis, and blood donors tend to have more type O individuals in them, so they may not be an ideal comparison for people who have contracted COVID-19. And this bears out in our data: from the Manhattan plot in Figure 1, you can see we do not see a statistically significant result (i.e., points rising above the red line) above chromosome 9. This means that the COVID-19 HGI analysis, which includes data from the study in the NEJM, does not support the association of the ABO blood group gene at this stage. We need larger sample sizes to clarify whether this region is associated with COVID-19.
Acknowledging the limitations of our study
No study design is perfect, and we’d like to highlight a few limitations of our research. First, the results described above are preliminary and from data submission in July of 2020. While we have enough samples to make some initial observations, larger samples sizes in future iterations will help us become confident in our conclusions. While larger sample sizes regrettably mean that more people have become infected with SARS-CoV-2, it also improves our ability to find patterns between host genetics and disease outcomes.
Second, the definition of disease severity can vary from one individual study to another. Furthermore, the controls are presumed to not have COVID-19, but we know that there are many asymptomatic individuals present in communities, so some of these “controls” may actually have contracted COVID-19. However, these limitations can be overcome by increasing the number of cases and controls assessed: the more samples we analyze, the lower the risk of observing a false positive signal due to the study design limitations. And once a positive signal is identified, we can focus on a smaller study with more specific definitions for our case and control groups to validate the finding. Ultimately, using our genetic findings to gain insights into the disease mechanisms requires additional research.
Our next steps
In order to address the limitation of sample size, we are continuing to accept submissions from contributing studies. The next analysis will be performed at the end of September and results will be released early October 2020. We hope to gain more insights in the next release of results, which promises to have a sample size of up to 50% the size that we have now. We also expect to collect richer data with more details about COVID-19 patient symptoms. Check back here to read about what we have learned in October 2020!
Using our preliminary results, the detective work begins. Our consortium and other scientists can perform additional studies to better understand the biological processes affected by these genes, and how that might be relevant for COVID-19 outcomes. If you would like more details on the follow up studies, head to this link. One such study will explore how this genetic variation is associated with outcomes specifically among the most severely affected hospitalized patients. We’re excited to understand our genetic findings further in the hopes it could lead to better clinical management of COVID-19 patients or disease treatments.
Further resources
To read more about COVID-19 Host Genetics Initiative, check out coverage in the popular press.
Acknowledgements
Thank you to Rachel Liao, Caitlin Cooney, CGC, Karen Zusi, Andrea Ganna, Alina Chan, Sophie Limou, Shea Andrews, and Jamal Nasir for thoughtful feedback and revisions.
Footnote
A Manhattan plot (aptly named because the peaks should look like the New York City skyline), is a common visualization of GWAS results. The horizontal line or x-axis (“Chromosome”) displays the positions of genetics variants across the 23 chromosomes (humans have 22 pairs of chromosomes plus some combination of the X and Y sex chromosomes). The vertical line or y-axis displays a measure of statistical significance called p-values, transformed to be in negative logarithmic scale. Each point on the plot displays the statistical significance (the p-value) of the association between a genetic variant at a given chromosomal position (called a SNP, pronounced “snip”) with the disease outcome being measured in each person. The higher the point is on the vertical axis, the more likely it is that this SNP is associated with the outcome of interest (e.g., COVID-19 severity). Our methodology is cautious: whereas many studies require a p-value less than 0.05 to consider a finding to be significant, we require a p-value less than 0.00000005 (indicated by the red line) to improve the confidence in our findings. If the point is higher than the red line, we consider the genetic association to be “statistically significant” and therefore, can design experiments to further validate and to understand the SNPs biological relevance
