Exploring the puzzle of HLA immunogenetics in COVID-19 patients
December 15, 2021
Written by Venceslas Douillard, Jamal Nasir, PhD, Nathan Pearson, PhD, Jeffrey Rosenfeld, PhD and Sophie Limou, PhD
Edited by Brooke Wolford, PhD
Note: This blog is intended for an audience of scientists and contains terminology specific to the field of genetics & genomics.
The emergence of SARS-CoV-2 shook our lives almost two years ago. The virus has infected at least 251 million individuals and caused more than 5 million deaths globally (https://covid19.who.int/, November 2021). The wide variability observed in disease susceptibility, severity and prognosis suggests that host genetic factors play a key role in immune responses against the virus. The COVID-19 host genetics initiative (HGI) aims to characterize the genetic determinants of COVID-19 outcomes by bringing together human genetics experts from across the globe. For the last results of the COVID-19 HGI, please see this summary and our recent results update in medRxiv and Nature.
What is HLA?
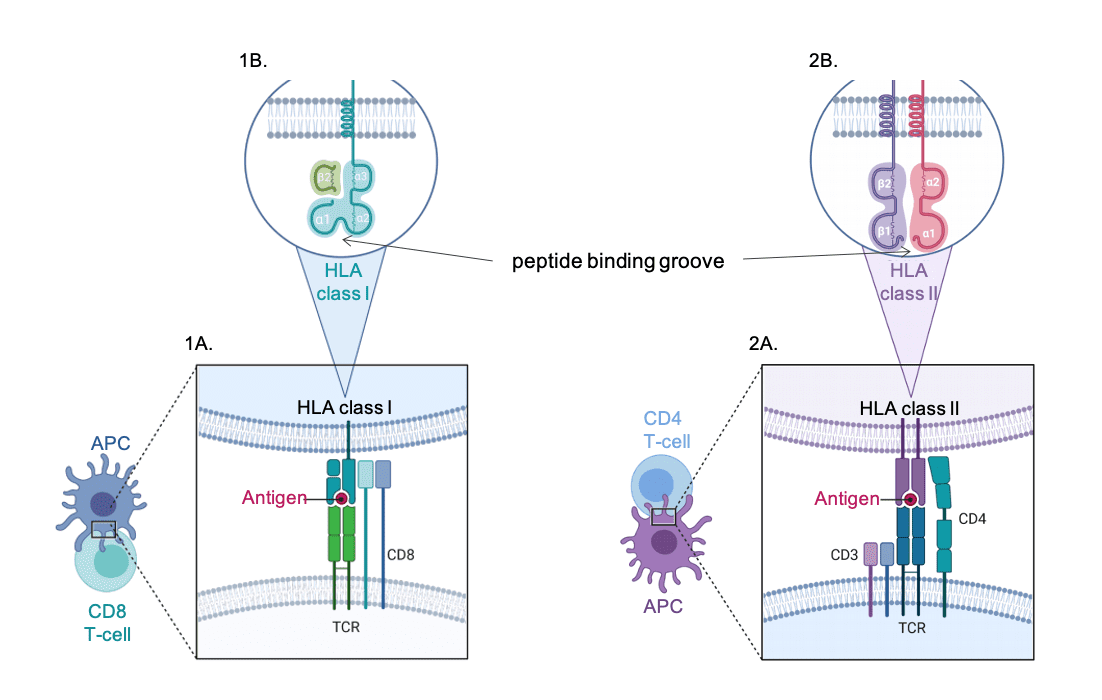
The classical Human Leukocyte Antigen (HLA) genes are located on chromosome 6 within the human major histocompatibility complex (MHC) that was initially investigated for its seminal role in transplantation outcomes. Classical HLA genes encode for cell surface molecules that process and present endogenous and exogenous antigens to the immune system. These molecules are essential for triggering efficient immune responses against non-self-peptides, including in the context of infectious diseases. Class I HLA molecules (e.g., A, B and C) are ubiquitously expressed and specifically interact with CD8 T cells, when class II HLA molecules (eg. DR, DQ, DP) are expressed by antigen-presenting cells and specifically activate CD4 T cells.
Figure 1 | HLA molecules and antigen presentation.
Left panel 1 presents class I HLA molecules and right panel 2 presents the class II HLA molecules.
A) Molecular interactions. B) HLA molecule structure. APC, antigen presenting cells.
Figure credit: Estelle Geffard, PhD
HLA genes exhibit an exceptional diversity with more than 27,000 alleles described for the 5 most polymorphic loci (A, B, C, DRB, DQB1) (https://www.ebi.ac.uk/ipd/imgt/hla/stats.html). Most of this diversity lies within the peptide binding groove, which allows peptide presentation of various shapes and sizes. This feature guarantees a variety of immune responses against pathogens at a populational scale and reflects the ongoing evolutionary arms race between pathogens and human populations across the globe.
Why is HLA important in infectious diseases?
The HLA region is known as a major determinant for disease risk, with thousands of genetic associations reported in infectious diseases, autoimmunity, cancers, and other traits. HLA associations have been reported with HLA alleles in focused immunogenetic studies, as well as with SNPs from the HLA loci. Indeed, 2.5% of the significant associations reported in the GWAS catalog map in the extended HLA/MHC region, and 21% of traits reported in this catalog exhibit at least one association in the extended HLA region.
Amongst the numerous associations reported in infectious diseases (Dendrou et al. Nat Rev Immunol 2018, Sanchez-Mazas Swiss Med Week 2020), HLA-B57 alleles and tagging SNPs were strongly associated with HIV-1 (Human Immunodeficiency Virus) viral control in African (HLA-B57:03 tagged by the rs2523608 SNP) and European (HLA-B57:01 tagged by the rs2395029 SNP) ancestry populations (Pelak et al. JID 2010, Fellay et al. Science 2007, Limou et al. JID 2009). A SNP located 35kb upstream HLA-C and impacting HLA-C surface expression levels was also associated with HIV control (International HIV Controllers Study et al. Science 2010, Thomas et al. Nat Genet 2009, Kulkarni et al. Nature 2011). Class II HLA alleles were associated with an increased risk for HBV (Hepatitis B Virus) infection and 3’UTR HLA-DPA1 and HLA-DPB1 SNPs were found to confer protection against chronic HBV in Asians (Jiang et al. Hepatology 2015, Hu et al. Nat Genet 2013). Because of its pivotal role in the immune response to pathogens, we suspect that HLA variation could also play a role in SARS-CoV-2 infection and/or COVID-19 outcomes.*
Why is it challenging to study the HLA region?
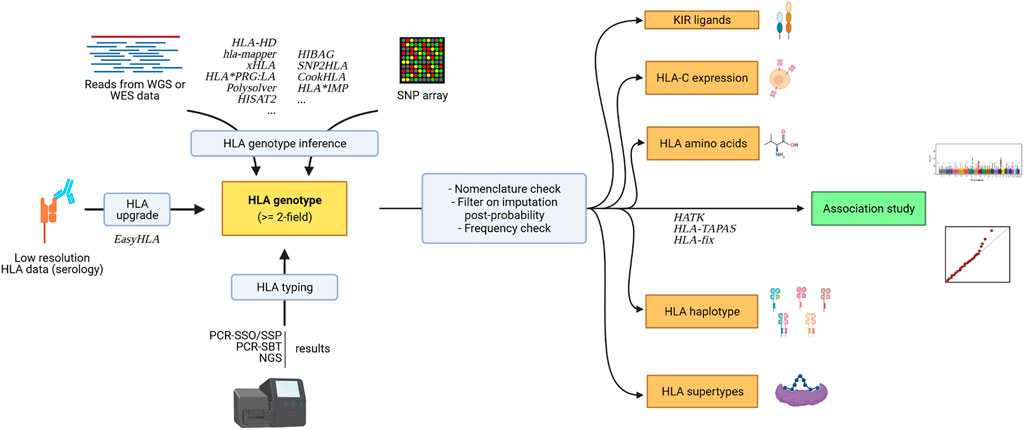
The genetic diversity and complexity of the whole MHC locus warrant the implementation of specific tools and strategies to appropriately analyze the role of HLA alleles against SARS-CoV-2 and COVID-19 phenotypes (for a recent review on HLA analytical tools and methods, see Douillard et al. Front Genet 2021). Over the past decades, the technologies to generate HLA data have evolved from serology-based techniques to next-generation sequencing. The HLA resolution and nomenclature have evolved to a complex description of the allele diversity: from a simple serotype (e.g., HLA-A02), to the protein sequence description (e.g., HLA-A02:01* and HLA-A02:01:01* for non-synonymous and synonymous variations, respectively) and to non-coding variants (e.g., HLA-A02:01:01:134). Beyond direct HLA typing, high-density SNP genotyping microarrays have empowered genome-wide association studies (GWAS) and numerous SNPs from the extended MHC region have thus been associated with immune-related traits. However, SNPs are simple genetic markers that do not capture the complexity of linkage disequilibrium patterns and the functional molecular mechanisms of HLA alleles. Powerful SNP to HLA imputation methods have recently been developed which propose a statistical prediction of HLA alleles from GWAS SNP datasets using reference population panels (with both HLA allele and SNP datasets) and machine learning algorithms (Dilthey et al. Bioinformatics 2011, Zheng et al. Pharmacogenomics J 2014, Cook et al. Nat Comm 2021, Naito et al. Nat Comm 2021). The quality of HLA imputation is highly dependent on informative matching reference panels, and the international SNP-HLA reference consortium (SHLARC) has recently been launched to promote, improve and facilitate HLA allele inference from GWAS SNPs (Vince et al. Genet Epidemiol 2020). Interestingly, statistical inference methods can also be implemented to generate additional layers of complex functional immunogenetic parameters (e.g., HLA amino acids, 5-gene HLA haplotype, interactions with KIR molecules, cell surface expression) (Geffard et al. Bioinformatics 2020).
Figure 2 | Generation of HLA data for immunogenetic-driven association studies.
The name of several HLA-dedicated tools are indicated in italic.
Figure credit: Venceslas Douillard (see Douillard et al Front Genet 2021)
Another challenge for HLA association studies pertains to the particular distribution of HLA allele frequencies. Indeed, the very high number of HLA alleles is associated to low or very low allelic frequency, which limits statistical power for signal detection and requires very large study population.
Does HLA contribute to COVID-19 outcomes?
The first HLA-centric studies performed on SARS-CoV-2 infection and/or COVID-19 severity explored the correlations between HLA allele frequencies in population databases and COVID-19 incidence, but they failed to identify significant associations or revealed signals that were discordant between studies (Pisanti et al. J Trans Med 2020, Tomita et al. Immun Infl Dis 2020, Toyoshima et al. J Hum Genet 2020).
The association studies of direct HLA typing with COVID-19 outcomes mostly reported inconclusive or unreproducible results due to the small sample sizes (n=45 to 265 individuals), difference in genetic ancestries (e.g., Brazil, China, Italy, Russia, Spain), and divergence in phenotype definitions (e.g., positive viral test, reported COVID-19, hospitalized cases) (Iturrieta-Zuazo et al. Clin Immunol 2020, Lorente et al. Med Intensiva 2021, Wang et al. HLA 2020, Novelli et al. HLA 2020, Yung et al. HLA 2020, Khor et al. Front Immunol 2021, Amoroso et al. Transplantation 2021, Castelli et al. Front Immunol 2021, Anzurez et al. HLA 2021, Shkurnikov et al. Front Immunol 2021). As examples, the study of 190 Chinese patients with confirmed COVID-19 revealed an association between the B22 serotype and SARS-CoV-2 infection (P=0.032, OR=1.71)(Yung et al. HLA 2020), when the study of 190 Japanese patients with mild to severe COVID-19 identified an association for HLA-A11:01* with COVID-19 severity (P=0.013, OR=2.26)(Khor et al. Front Immunol 2021). Importantly, the study of known HLA genotypes in close to 73,000 Israeli individuals, including 6,413 who tested positive for SARS-CoV-2 infection and 181 who were hospitalized for COVID-19, failed to identified any significant association with common HLA alleles (Shachar et al. JCI 2021).
The inference of HLA alleles from next-generation sequencing (NGS) data in 332 hospitalized Chinese patients revealed frequency differences between severe and mild COVID-19 cases for HLA-C14:02* (P=0.003, OR=4.7), HLA-B51:01* (P=0.007, OR=3.3), and HLA-A11:01* (P=0.009, OR=3.3)(Wang et al. Cell Discov 2020).
Finally, the GWAS results from large populations, including the ones from the COVID-19 HGI meta-analysis, did not reveal a major role for HLA SNPs in SARS-CoV2 infection so far, but reported associations with COVID-19 severity (Ellinghaus et al. NEJM 2020, Covid-19 HGI Nature 2021, Covid-19 HGI MedRxiv 2021). Some heterogeneity has been observed between the cohorts, which might stem from variability in HLA allele frequency distribution or linkage disequilibrium patterns within the different populations enrolled in the meta-analysis (European, Admixed American, African, Middle Eastern, South Asian and East Asian individuals). The last COVID-19 HGI release (n=8,779 critical COVID-19 cases vs. 1,001,875 general population controls) reported 5 MHC variants reaching genome-wide statistical significance within the CCHCR1 gene, located 110kb downstream of HLA-C (top SNP rs111837807, P=2.2x10-11, OR=1.2), as well as a variant within HLA-DPB1 3’UTR (rs9501257, P=4.1x10-8, OR=1.2). These results suggest that further increasing cohort sizes from each ancestry and/or running in-depth HLA-centric explorations within large cohorts will better elucidate the role of HLA immunogenetic factors in COVID-19 outcomes.
Conclusions
So far, genomic and immunogenic investigations have revealed a milder impact for HLA molecules in the SARS-CoV-2 pandemic than what could have been anticipated based on infectious diseases such as HIV. However, the precise role of HLA molecules is yet to be fully defined and HLA-specific studies are warranted. The global effort of the international scientific community in large genomic cohorts should be a springboard for HLA-centric immunogenomic analyses by leveraging GWAS SNP and NGS data with HLA imputation and with further immunogenetic parameters statistical inference (e.g., 5-gene haplotypes). The HLA COVID-19 HLA & Immunogenetics Consortium (CHIC, http://www.hlacovid19.org/) will support such collective initiatives by providing an analysis platform, expertise, and a database to describe the role of HLA complex molecules in COVID-19 outcomes.