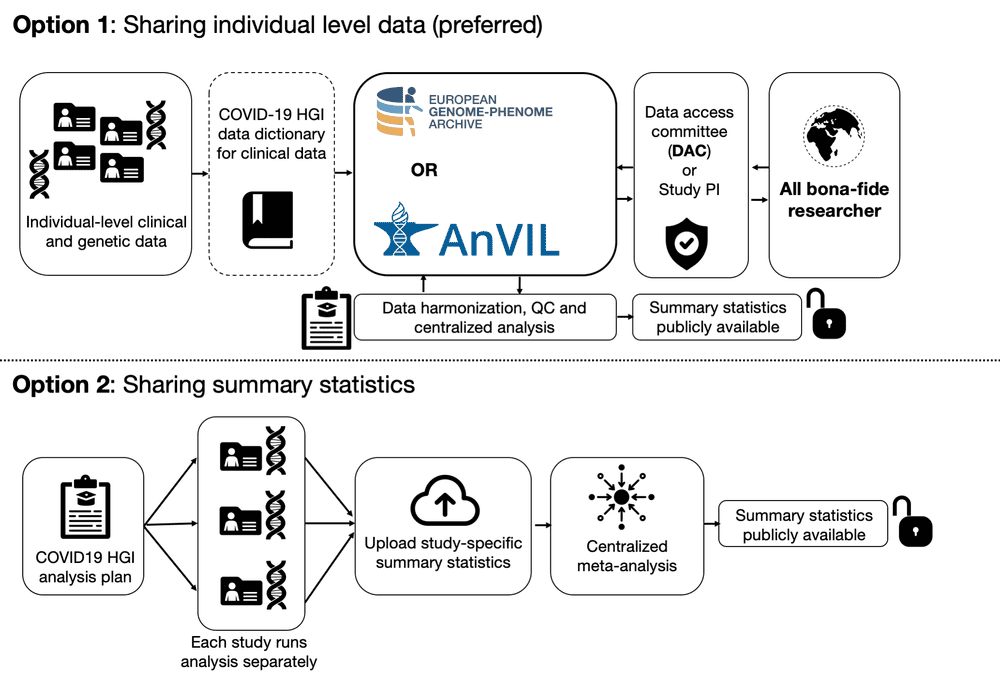
How can I submit my data?
You can submit data in two ways. You can submit results summary statistics (calculated and formatted according to the analysis plan) or you can submit individual-level data.
We prefer you submit individual-level data because they can be used beyond the few analyses that are described in the analysis plan.
Results summary statistics
Information on how to upload results summary statistics are given in the analysis plan in the section “Results upload instructions”
Individual-level data
If you are not from US:
- You can submit individual-level data (i.e. genetic and clinical phenotype data) via the European Genome-phenome Archive (EGA). EGA offers services for archiving, processing and distribution for all types of potentially identifiable genetic and phenotypic human data at the European Bioinformatics Institute (EBI). To start your submission please fill this form or contact the EGA helpdesk via helpdesk@ega-archive.org and mark the email F.A.O Giselle Kerry stating that your submission is part of the COVID-19 Host Genetics Initiative.
If you are from the US:
- You can submit individual-level data via NHGRI AnVIL. The AnVIL can ingest datasets, process them via standardized pipelines and perform quality control on them, and make them accessible to other researchers in a cloud-based environment. To start your submission, please contact COVID@lists.anvilproject.org and mark the email Attn: COVID-19 Host Genetics Initiative.
Where can I access individual-level data?
A small subset of individual-level data and clinical phenotypes that have been analyzed as part of the COVID-19 Host Genetics Initiative are made avialable via EGA HERE
Where can I access results summary statistics?
Results summary statistics are meta-analyzed across studies and immediately made available to the scientific community via the website result browser, via GWAS catalog, Open Target Platform and other portals.
Can I access summary statistics from the individual studies?
On the result page, we make available the meta-analysis summary statistics for the combined studies. We also make available leave-one-out analysis. However, to access the study-specific summary statistics you will need to get in contact with each study PI separately.
My data cannot leave my country, what can I do?
The EGA is working with the ELIXIR network to establish the EGA Federation network to enable data to be deposited within national jurisdictions. We expect to launch the first nodes in mid-late 2020. In the meantime, we suggest you contact your country's ELIXIR head of node to find out about the current status for your country.
In which format should I submit the genetic data?
Both EGA and AnVIL recommend using open standards and formats that are maintained by the Global Alliance for Genomics and Health (GA4GH), published in the GA4GH Genomic Data Toolkit. For genome sequencing data this includes FASTQ, BAM, CRAM, and VCF. All array-based technologies are accepted, which may include the raw data, intensity and analysis files, and there are no restrictions on data formats accepted.
Is the system compliant with current European regulations for data protection?
The EGA is managed by EMBL-EBI and Center for Genome Regulation, Barcelona (CRG). At EMBL, that protection is enacted by the Internal Policy 68 on general data protection (IP 68). IP 68 resembles the GDPR, but adapts to the intergovernmental nature of EMBL and to the needs of enabling free scientific research across national borders. CRG is subject to the GDPR and implements it fully. The EGA GDPR notices can be found here.