Release Date
July 2, 2020
Study Abbreviations
| Full Name | Abbreviation |
|---|---|
| Genetic determinants of COVID-19 complications in the Brazilian population | BRACOVID |
| Genetic modifiers for COVID-19 related illness | BelCovid |
| deCODE | DECODE |
| FinnGen | FinnGen |
| GEN-COVID, reCOVID | GENCOVID |
| UK 100,000 Genomes Project | genomicsengland100kgp_EUR |
| Genes & Health | GNH |
| Generation Scotland | GS |
| COVID19-Host(a)ge | HOSTAGE |
| Helix Exome+ COVID-19 Phenotypes | Helix |
| UK Blood Donors Cohort | INTERVAL |
| LifeLines CytoSNP | LifelinesCyto |
| LifeLines Global Screening Array | LifelinesGsa |
| Netherlands Twin Register | NTR |
| Partners Healthcare Biobank | PHBB |
| Qatar Genome Program | QGP |
| UK Biobank | UKBB |
Data Columns
| Column Name | Description |
|---|---|
| #CHR | chromosome |
| POS | chromosome position in build 37 |
| REF | non-effect allele |
| ALT | effect allele (beta is for this allele) |
| SNP | #CHR:POS:REF:ALT |
| {STUDY}_AF_Allele2 | allele frequency in {STUDY} |
| {STUDY}_AF_fc | allele frequency in {STUDY} / allele frequency in gnomAD v3 (1000000 if frequency in gnomAD is 0). Calculated based on each study's ancestry in gnomAD |
| all_meta_N | number of studies that had the variant after AF and INFO filtering and as such were used for the meta |
| all_inv_var_meta_beta | effect size on log(OR) scale |
| all_inv_var_meta_sebeta | standard error of effect size |
| all_inv_var_meta_p | p-value |
| all_inv_var_het_p | p-value from Cochran's Q heterogeneity test |
Release Notes
Meta-analysis was done with inverse variance weighting. Analysis carried out on GRCh38; results also available through GRCh37 liftover. An AF filter of 0.0001 and an INFO filter of 0.6 was applied to each study prior to meta-analysis.
ANA_A2_V2
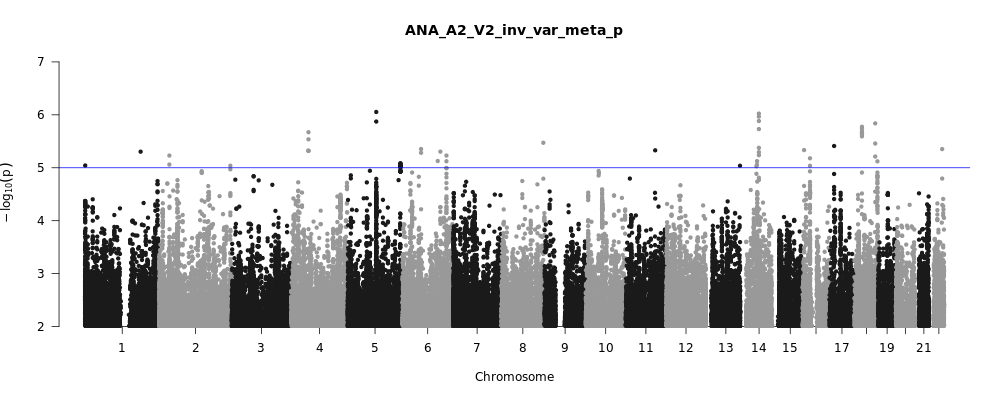
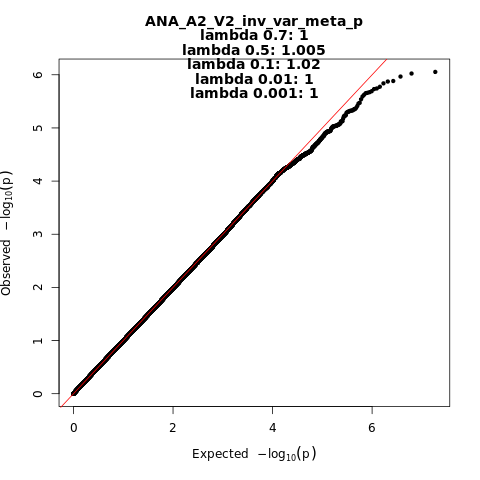
Phenotype
very severe respiratory confirmed covid vs. population
Population
All
Total Cases
536
Total Controls
329391
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| GENCOVID_EUR | 327 | 2461 |
| UKBB_EUR | 108 | 325397 |
| BRACOVID_AMR | 101 | 1533 |
Downloads
GRCh37 liftover: COVID19_HGI_ANA_A2_V2_20200701.b37.txt.gz
GRCh37 liftover (.tbi): COVID19_HGI_ANA_A2_V2_20200701.b37.txt.gz.tbi
GRCh37 liftover (filtered): COVID19_HGI_ANA_A2_V2_20200701.b37_1.0E-5.txt
GRCh38: COVID19_HGI_ANA_A2_V2_20200701.txt.gz
GRCh38 (.tbi): COVID19_HGI_ANA_A2_V2_20200701.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_ANA_A2_V2_20200701.txt.gz_1.0E-5.txt
ANA_B1_V2
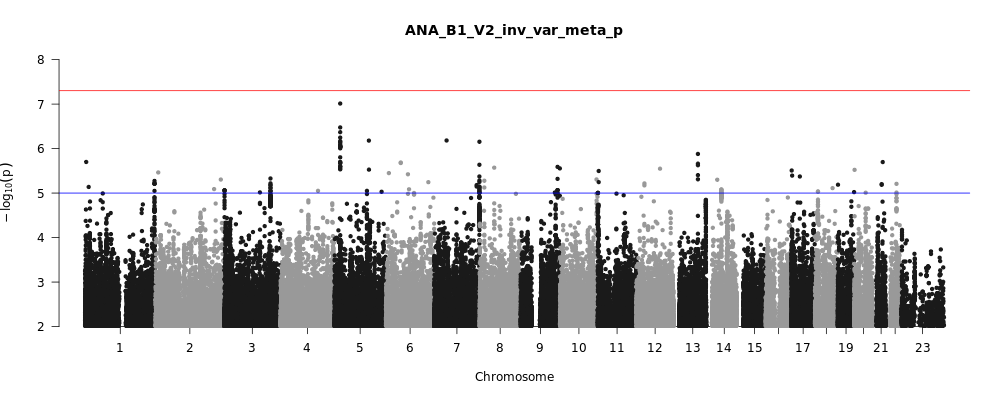
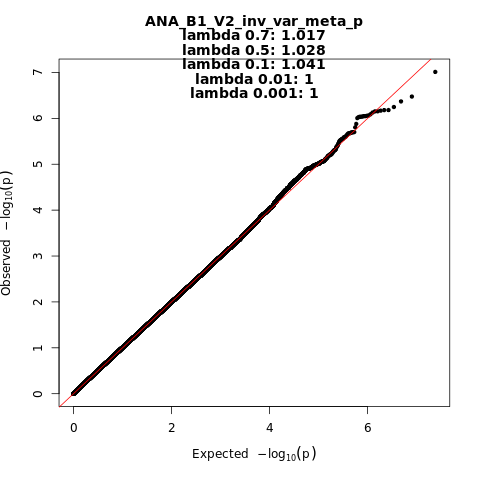
Phenotype
hospitalized covid vs. not hospitalized covid
Population
All
Total Cases
928
Total Controls
2028
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| UKBB_EUR | 784 | 406 |
| DECODE_EUR | 86 | 1462 |
| FinnGen_FIN | 58 | 160 |
Downloads
GRCh37 liftover: COVID19_HGI_ANA_B1_V2_20200701.b37.txt.gz
GRCh37 liftover (.tbi): COVID19_HGI_ANA_B1_V2_20200701.b37.txt.gz.tbi
GRCh37 liftover (filtered): COVID19_HGI_ANA_B1_V2_20200701.b37_1.0E-5.txt
GRCh38: COVID19_HGI_ANA_B1_V2_20200701.txt.gz
GRCh38 (.tbi): COVID19_HGI_ANA_B1_V2_20200701.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_ANA_B1_V2_20200701.txt.gz_1.0E-5.txt
ANA_B2_V2
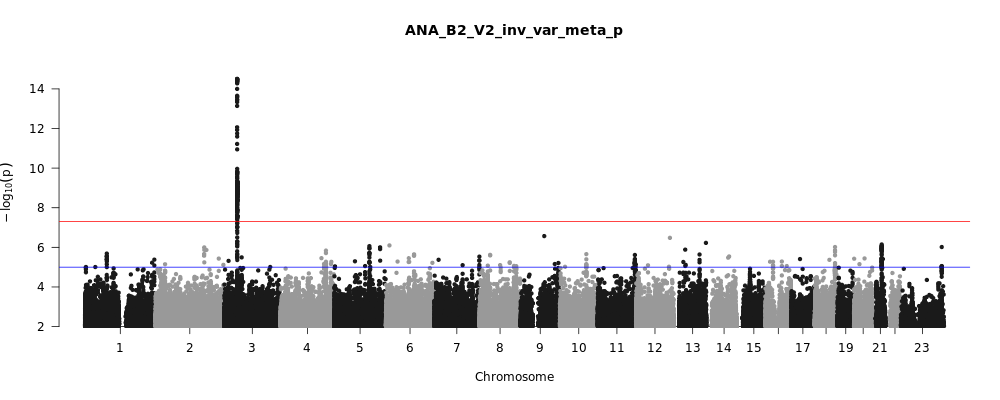
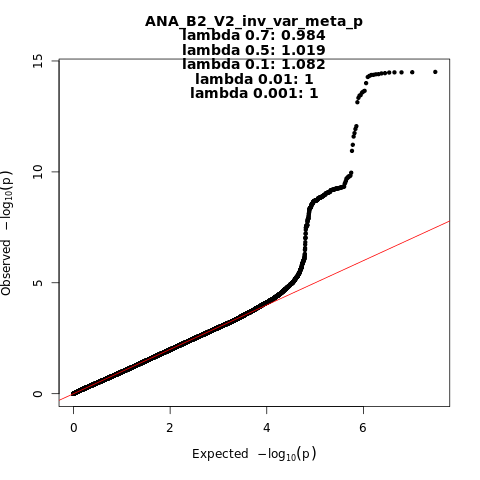
Phenotype
hospitalized covid vs. population
Population
All
Total Cases
3199
Total Controls
897488
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| HOSTAGE_EUR | 1610 | 2205 |
| UKBB_EUR | 784 | 419747 |
| GENCOVID_EUR | 366 | 2461 |
| BRACOVID_AMR | 176 | 1533 |
| DECODE_EUR | 86 | 223971 |
| GNH_SAS | 62 | 27353 |
| FinnGen_FIN | 58 | 218734 |
| BelCovid_EUR | 57 | 1484 |
Downloads
GRCh37 liftover: COVID19_HGI_ANA_B2_V2_20200701.b37.txt.gz
GRCh37 liftover (.tbi): COVID19_HGI_ANA_B2_V2_20200701.b37.txt.gz.tbi
GRCh37 liftover (filtered): COVID19_HGI_ANA_B2_V2_20200701.b37_1.0E-5.txt
GRCh38: COVID19_HGI_ANA_B2_V2_20200701.txt.gz
GRCh38 (.tbi): COVID19_HGI_ANA_B2_V2_20200701.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_ANA_B2_V2_20200701.txt.gz_1.0E-5.txt
ANA_C1_V2
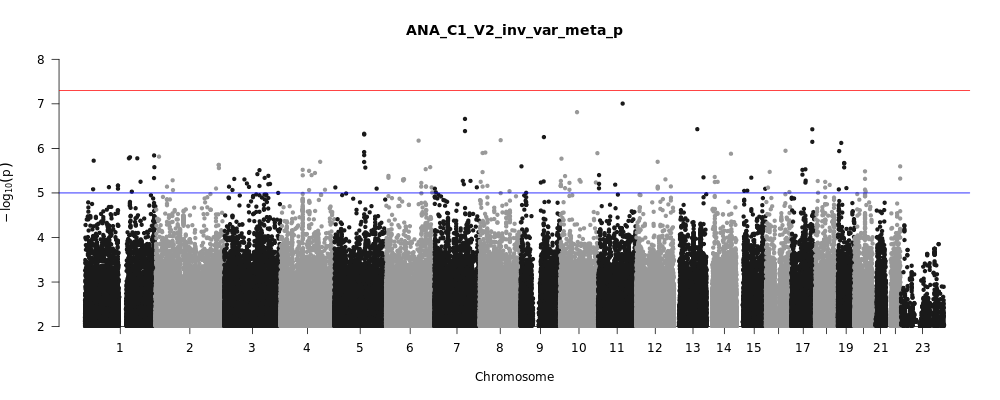
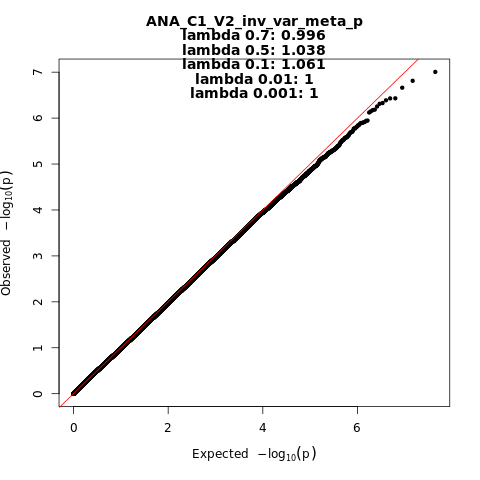
Phenotype
covid vs. lab/self-reported negative
Population
All
Total Cases
3523
Total Controls
36634
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| DECODE_EUR | 1597 | 29065 |
| UKBB_EUR | 1190 | 5014 |
| genomicsengland100kgp_EUR | 147 | 562 |
| INTERVAL_EUR | 137 | 472 |
| NTR_EUR | 121 | 108 |
| GNH_SAS | 114 | 256 |
| PHBB_EUR | 97 | 904 |
| UKBB_AFR | 63 | 125 |
| UKBB_CSA | 57 | 128 |
Downloads
GRCh37 liftover: COVID19_HGI_ANA_C1_V2_20200701.b37.txt.gz
GRCh37 liftover (.tbi): COVID19_HGI_ANA_C1_V2_20200701.b37.txt.gz.tbi
GRCh37 liftover (filtered): COVID19_HGI_ANA_C1_V2_20200701.b37_1.0E-5.txt
GRCh38: COVID19_HGI_ANA_C1_V2_20200701.txt.gz
GRCh38 (.tbi): COVID19_HGI_ANA_C1_V2_20200701.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_ANA_C1_V2_20200701.txt.gz_1.0E-5.txt
ANA_C2_V2
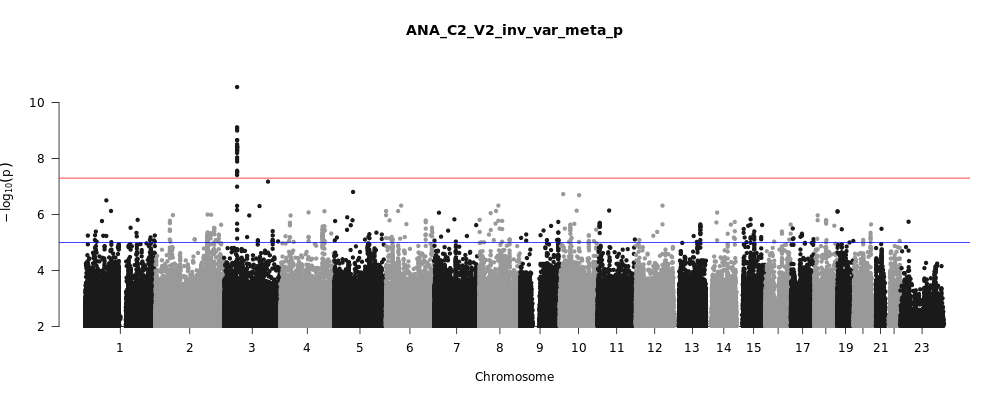
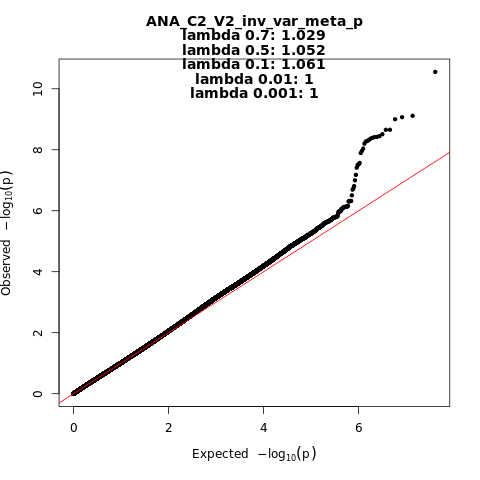
Phenotype
covid vs. population
Population
All
Total Cases
6696
Total Controls
1073072
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| HOSTAGE_EUR | 1610 | 2205 |
| DECODE_EUR | 1597 | 282150 |
| UKBB_EUR | 1190 | 419341 |
| GENCOVID_EUR | 407 | 2461 |
| FinnGen_FIN | 264 | 203376 |
| LifelinesGsa_EUR | 247 | 18403 |
| QGP_ARAB | 197 | 13698 |
| BRACOVID_AMR | 176 | 1533 |
| genomicsengland100kgp_EUR | 147 | 562 |
| INTERVAL_EUR | 137 | 41699 |
| Helix_EUR | 133 | 3404 |
| NTR_EUR | 121 | 4648 |
| GNH_SAS | 114 | 27301 |
| PHBB_EUR | 97 | 28927 |
| LifelinesCyto_EUR | 82 | 6488 |
| UKBB_AFR | 63 | 6573 |
| UKBB_CSA | 57 | 8819 |
| BelCovid_EUR | 57 | 1484 |
Downloads
GRCh37 liftover: COVID19_HGI_ANA_C2_V2_20200701.b37.txt.gz
GRCh37 liftover (.tbi): COVID19_HGI_ANA_C2_V2_20200701.b37.txt.gz.tbi
GRCh37 liftover (filtered): COVID19_HGI_ANA_C2_V2_20200701.b37_1.0E-5.txt
GRCh38: COVID19_HGI_ANA_C2_V2_20200701.txt.gz
GRCh38 (.tbi): COVID19_HGI_ANA_C2_V2_20200701.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_ANA_C2_V2_20200701.txt.gz_1.0E-5.txt
ANA_D1_V2
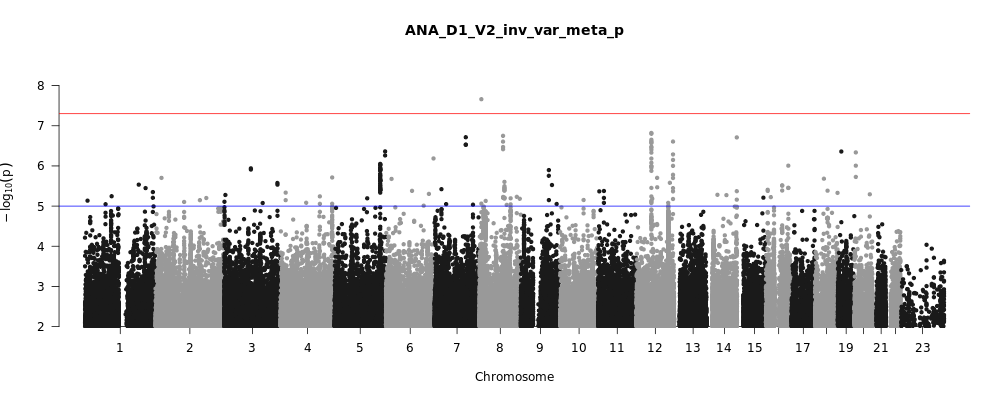
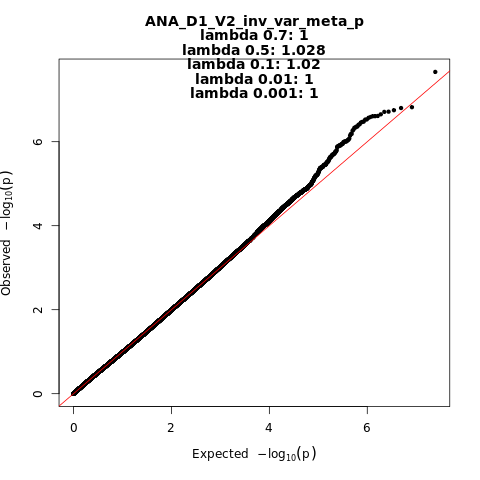
Phenotype
predicted covid from self-reported symptoms vs. predicted or self-reported non-covid
Population
All
Total Cases
1865
Total Controls
29174
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| NTR_EUR | 695 | 4284 |
| LifelinesGsa_EUR | 458 | 12836 |
| Helix_EUR | 398 | 3959 |
| LifelinesCyto_EUR | 182 | 4485 |
| GS_EUR | 132 | 3610 |
Downloads
GRCh37 liftover: COVID19_HGI_ANA_D1_V2_20200701.b37.txt.gz
GRCh37 liftover (.tbi): COVID19_HGI_ANA_D1_V2_20200701.b37.txt.gz.tbi
GRCh37 liftover (filtered): COVID19_HGI_ANA_D1_V2_20200701.b37_1.0E-5.txt
GRCh38: COVID19_HGI_ANA_D1_V2_20200701.txt.gz
GRCh38 (.tbi): COVID19_HGI_ANA_D1_V2_20200701.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_ANA_D1_V2_20200701.txt.gz_1.0E-5.txt
