Release Date
April 8, 2022
Study Abbreviations
| Full Name | Abbreviation |
|---|---|
| 23andMe | 23ANDME |
| 24Genetics | idipaz24genetics |
| AArmenia_Covid-19hg | ArmCovid |
| ACCOuNT | ACCOuNT |
| Adolescent Brain and Cognitive development study | ABCD |
| Amsterdam UMC COVID study group | Amsterdam_UMC_COVID_study_group |
| Ancestry Freeze Four | ANCESTRY_Freeze_Four |
| Assessment of the influence of clinical, functional, immunological and genetic factors on the severity of the course of coronavirus infection with SARS-CoV-2 and Post Covid syndrome | KazCovid |
| Avon Longitudinal Study of Parents and Children (ALSPAC) | ALSPAC_G1 |
| Biobanque Quebec COVID19 | BQC19 |
| Bonn Study of COVID19 genetics | BoSCO |
| CCHC COVID-19 GAWS | CCHC |
| CGEn HostSeq - Canadian COVID-19 Human Host Genome Sequencing Databank | CGEN |
| CHOP CAG | CHOP_CAG |
| Columbia University COVID19 Biobank | CU |
| COMRI/Virology Study | COMRI |
| Coronagenes | Coronagenes |
| Covid19 Ioannina Biobank | Ioannina |
| COVID19-Host(a)ge | HOSTAGE |
| Covid19hg-CL | ChiCovid |
| deCODE | DECODE |
| Determining the Molecular Pathways and Genetic Predisposition of the Acute Inflammatory Process Caused by SARS-CoV-2 | SPGRX |
| Egypt hgCOVID hub | Egypt_hgCOVID_hub |
| EraCORE | ERACORE |
| Estonian Biobank | EstBB |
| EXCEED | EXCEED |
| FHoGID | FHoGID |
| FinnGen | FinnGen |
| French hgCOVID | FrenchCovid |
| Geisinger Health System | GHS_Freeze_145 |
| GEN-COVID, reCOVID | GENCOVID |
| Generation Scotland | Generation_Scotland |
| Genes & Health | GNH |
| Genes for Good | GFG |
| Genetic determinants of COVID-19 complications in the Brazilian population | BRACOVID |
| Genetic influences on severity of COVID-19 illness in Korea | Genetics_COVID19_Korea |
| Genetic modifiers for COVID-19 related illness | BelCovid |
| Genetics of COVID-related Manifestation | Corea |
| Genomes for Life | GCAT |
| Genomic epidemiology of SARS-Cov-2 and host genetics in Coronavirus Disease 2019 (COVID-19) | Stanford |
| genomiCC | genomicc |
| Genomics England | genomicsengland100kgp |
| Genotek COVID-19 study | Genotek |
| Helix Exome+ COVID-19 Phenotypes | Helix |
| Host genetic factors for COVID-19 severity and outcome in western Indian population | IND_GJ_COVID19 |
| Host genetic factors in COVID-19 patients in relation to disease susceptibility, disease severity and pharmacogenomics | thaicovid |
| Host Genetics in COVID cohorts of mixed ancestry from Mexico | MexCovid |
| HUNT | HUNT |
| Iran Covid | IranCovid |
| Japan Coronavirus Taskforce | JapanTaskForce |
| Jordan COVID-19 Host Genomics Initiative: (JCHGI) | JorCovid |
| Latvia COVID-19 research platform | LGDB |
| Lifelines | Lifelines |
| MexGen-COVID Initiative | MexGen-COVID |
| Michigan Genomics Initiative | MGI |
| Million Veterans Program | MVP |
| Mount Sinai Health System COVID-19 Genomics Initiative | SINAI_COVID |
| NCGM biobank | JSA-COVID19 |
| Netherlands Twin Register | NTR |
| Pa-COVID-19 | Charite |
| Partners Healthcare Biobank | PHBB |
| Penn Medicine Biobank | PMBB |
| Qatar Genome Program | QGP |
| SARS-CoV-2 and host genome sequencing | PakCovid |
| Saudi Human Genome Program | SaudiHumanGenomeProgram |
| Search for genomic markers predicting the severity of the response to COVID-19 | POLISH_COVID_WGS |
| Spanish COalition to Unlock Research on host GEnetics on COVID-19 (SCOURGE) | SCOURGE |
| The Colorado Center for Personalized Medicine | CCPM |
| The Danish Blood Donor Study | DBDS |
| The genetic predisposition to severe COVID-19 | SweCovid |
| The Norwegian Mother, Father and Child Cohort Study | MOBA |
| Tirschenreuth Study (TiKoco) | TIKOCO |
| TOPMed CHRIS | TOPMed_CHRIS10K |
| UCLA Precision Health COVID-19 Biobank | UCLA |
| UK Biobank | UKBB |
| UK Blood Donors Cohort | INTERVAL |
| Val Gardena | TOPMed_Gardena |
| Vanda COVID | Vanda |
| Vanderbilt Biobank | BioVU |
| Variability in immune response genes and severity of SARS-CoV-2 infection (INMUNGEN-CoV2 study) | INMUNGEN_CoV2 |
| Women's Health Genome Study | WGHS |
Data Columns
| Column Name | Description |
|---|---|
| #CHR | chromosome |
| POS | chromosome position |
| REF | reference and non-effect allele |
| ALT | alternative and effect allele (beta is for this allele) |
| SNP | #CHR:POS:REF:ALT |
| all_meta_N | number of studies that had the variant after AF 0.001 and INFO 0.6 filtering and as such were used for the meta |
| all_inv_var_meta_beta | effect size on log(OR) scale |
| all_inv_var_meta_sebeta | standard error of effect size |
| all_inv_var_meta_p | p-value |
| all_inv_var_meta_cases | total number of cases |
| all_inv_var_meta_controls | total number of controls |
| all_inv_var_meta_effective | effective sample size |
| all_inv_var_het_p | p-value from Cochran's Q heterogeneity test |
| lmso_inv_var_beta | leave-most-significant-study-out analysis effect size |
| lmso_inv_var_se | leave-most-significant-study-out analysis standard error of effect size |
| lmso_inv_var_pval | leave-most-significant-study-out analysis p-value |
| all_meta_AF | allele frequency in the meta-analysis |
| rsid | risd |
Release Notes
Meta-analysis was done with fixed effects inverse variance weighting. Results are available in genome builds 38 and 37. An AF filter of 0.001 and an INFO filter of 0.6 was applied to each study before meta.
Extras
The downloads below do not contain the 23andMe samples (the most well-powered analyses), however the top 10K SNPs from analyses that include 23andMe samples are available here.
There are additional results with links in this README: covidhgi-freeze-7-readme.txt.
Alternatively, you can browse the Google Cloud storage bucket: gs://covid19-hg-public/freeze_7/results/20220403. You will need gsutil to list and download these files.
Included at gs://covid19-hg-public/freeze_7/results/20220403/pop_spec/sumstats are ancestry-specific (AFR, EUR, HIS/AMR) and admixed (AFR/HIS/AMR) results for hospitalization and susceptibility scans. Also the top 10k variants including 23andMe, as well as leave UKBB out results for all ancestries and EUR only.
A2_ALL_leave_23andme
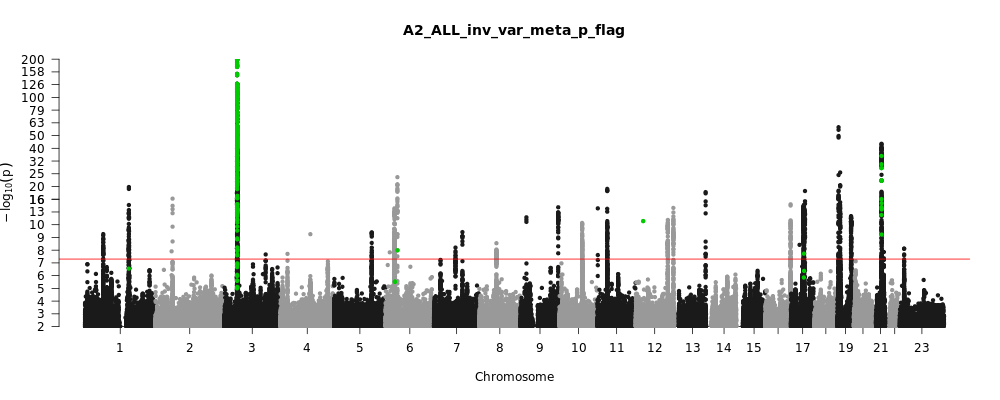
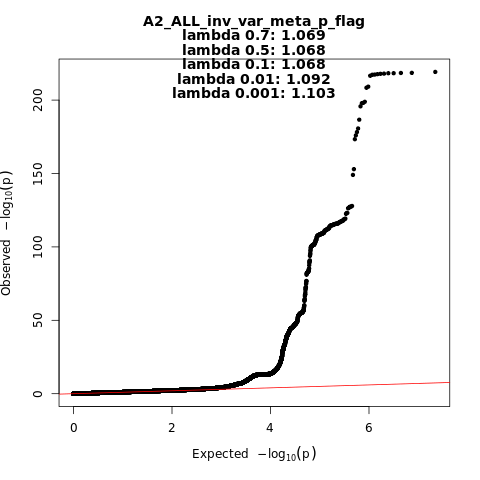
Phenotype
Very severe respiratory confirmed covid vs. population
Population
All
Total Cases
18152
Total Controls
1145546
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ANCESTRY_Freeze_Four_EUR | 667 | 113882 |
| Amsterdam_UMC_COVID_study_group_EUR | 66 | 1413 |
| BRACOVID_AMR | 539 | 1149 |
| CU_AFR | 133 | 2610 |
| CU_EUR | 203 | 2149 |
| FinnGen_FIN | 190 | 278193 |
| GENCOVID_EUR | 1077 | 2443 |
| GHS_Freeze_145_EUR | 321 | 108168 |
| PMBB_AFR | 55 | 8622 |
| SPGRX_EUR | 101 | 302 |
| SaudiHumanGenomeProgram_ARAB | 147 | 768 |
| Vanda_EUR | 58 | 900 |
| idipaz24genetics_EUR | 59 | 75 |
| BQC19_EUR | 202 | 1366 |
| BelCovid_EUR | 297 | 1477 |
| BioVU_EUR | 117 | 68064 |
| BoSCO_EUR | 195 | 984 |
| Charite_EUR | 87 | 865 |
| Egypt_hgCOVID_hub_MID | 699 | 472 |
| GNH_SAS | 192 | 43998 |
| HOSTAGE_EUR | 1850 | 12226 |
| INMUNGEN_CoV2_EUR | 81 | 723 |
| JapanTaskForce_EAS | 421 | 3317 |
| JorCovid_MID | 366 | 529 |
| MexGen-COVID_AMR | 230 | 5060 |
| POLISH_COVID_WGS_EUR | 224 | 1005 |
| SCOURGE_EUR | 1124 | 13224 |
| SweCovid_EUR | 181 | 3748 |
| UKBB_EUR | 680 | 419851 |
| genomicc_AFR | 440 | 1336 |
| genomicc_EAS | 274 | 352 |
| genomicc_EUR | 5989 | 41384 |
| genomicc_SAS | 788 | 3698 |
| JSA-COVID19_EAS | 99 | 1193 |
Downloads
GRCh37 liftover: COVID19_HGI_A2_ALL_leave_23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_A2_ALL_leave_23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_A2_ALL_leave_23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_A2_ALL_leave_23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_A2_ALL_leave_23andme_20220403_1e-5.tsv
A2_ALL_afr_leave_23andme
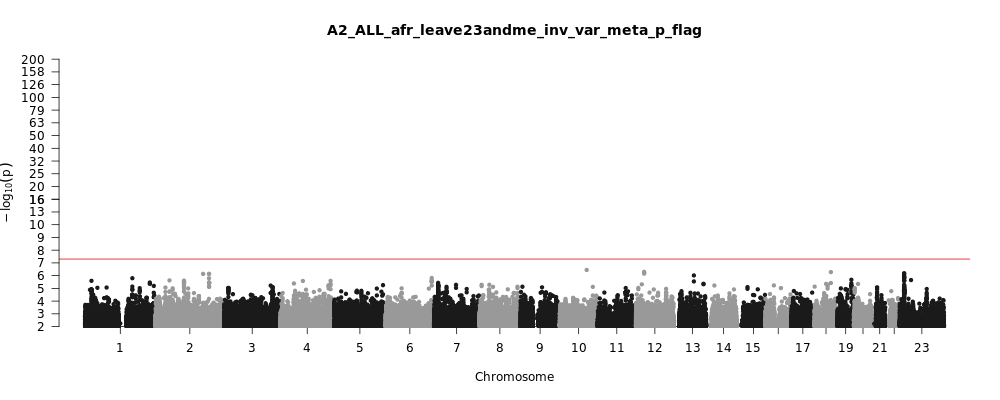
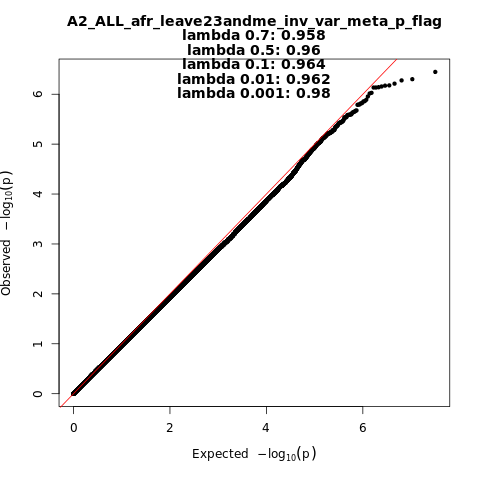
Phenotype
Very severe respiratory confirmed covid vs. population
Population
afr
Total Cases
628
Total Controls
12568
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| CU_AFR | 133 | 2610 |
| PMBB_AFR | 55 | 8622 |
| genomicc_AFR | 440 | 1336 |
Downloads
GRCh37 liftover: COVID19_HGI_A2_ALL_afr_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_A2_ALL_afr_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_A2_ALL_afr_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_A2_ALL_afr_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_A2_ALL_afr_leave23andme_20220403_1e-5.tsv
A2_ALL_eas_leave_23andme
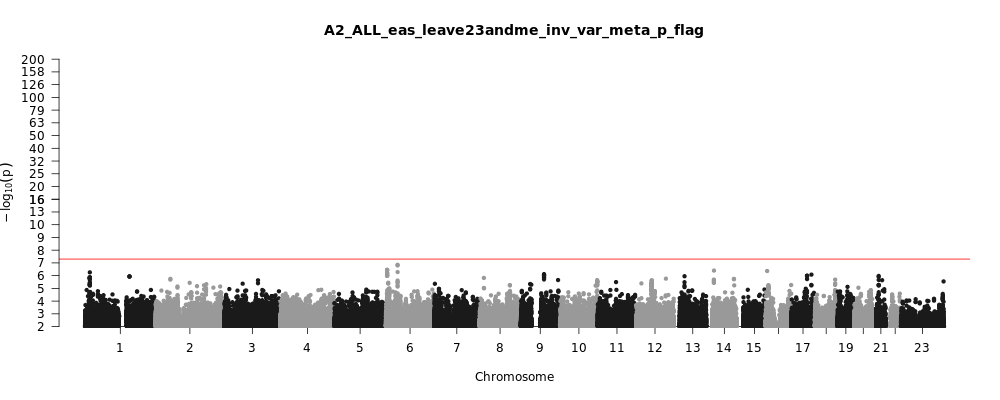
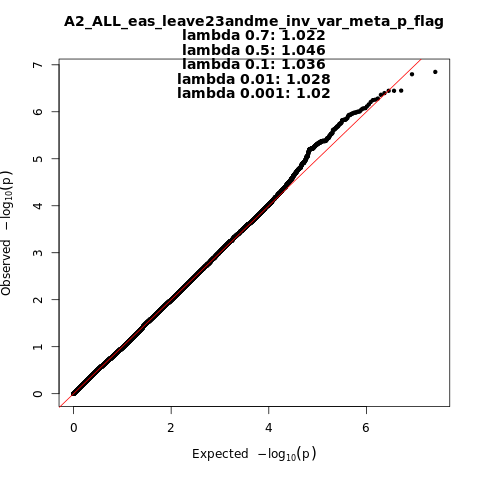
Phenotype
Very severe respiratory confirmed covid vs. population
Population
eas
Total Cases
794
Total Controls
4862
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| JapanTaskForce_EAS | 421 | 3317 |
| genomicc_EAS | 274 | 352 |
| JSA-COVID19_EAS | 99 | 1193 |
Downloads
GRCh37 liftover: COVID19_HGI_A2_ALL_eas_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_A2_ALL_eas_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_A2_ALL_eas_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_A2_ALL_eas_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_A2_ALL_eas_leave23andme_20220403_1e-5.tsv
A2_ALL_eur_leave_23andme
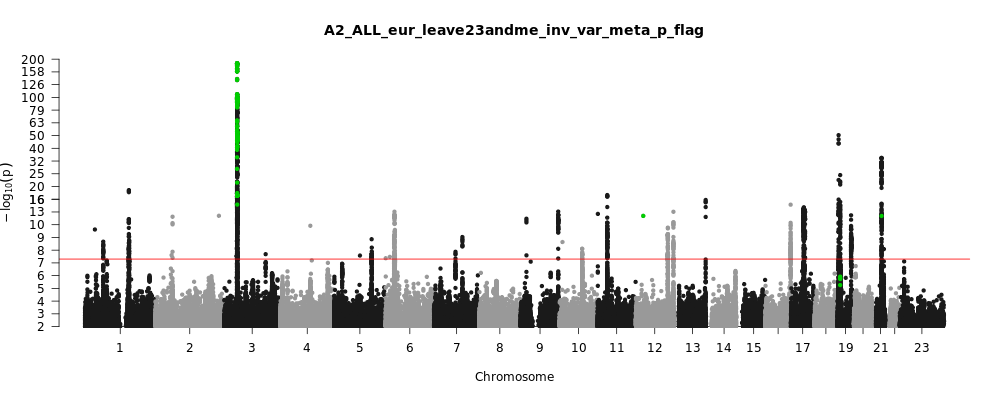
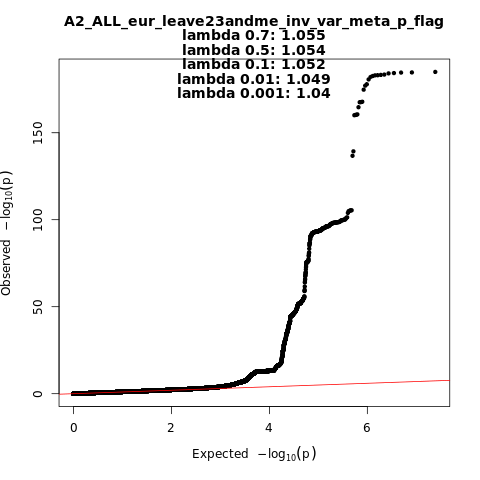
Phenotype
Very severe respiratory confirmed covid vs. population
Population
eur
Total Cases
13769
Total Controls
1072442
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ANCESTRY_Freeze_Four_EUR | 667 | 113882 |
| Amsterdam_UMC_COVID_study_group_EUR | 66 | 1413 |
| CU_EUR | 203 | 2149 |
| FinnGen_FIN | 190 | 278193 |
| GENCOVID_EUR | 1077 | 2443 |
| GHS_Freeze_145_EUR | 321 | 108168 |
| SPGRX_EUR | 101 | 302 |
| Vanda_EUR | 58 | 900 |
| idipaz24genetics_EUR | 59 | 75 |
| BQC19_EUR | 202 | 1366 |
| BelCovid_EUR | 297 | 1477 |
| BioVU_EUR | 117 | 68064 |
| BoSCO_EUR | 195 | 984 |
| Charite_EUR | 87 | 865 |
| HOSTAGE_EUR | 1850 | 12226 |
| INMUNGEN_CoV2_EUR | 81 | 723 |
| POLISH_COVID_WGS_EUR | 224 | 1005 |
| SCOURGE_EUR | 1124 | 13224 |
| SweCovid_EUR | 181 | 3748 |
| UKBB_EUR | 680 | 419851 |
| genomicc_EUR | 5989 | 41384 |
Downloads
GRCh37 liftover: COVID19_HGI_A2_ALL_eur_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_A2_ALL_eur_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_A2_ALL_eur_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_A2_ALL_eur_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_A2_ALL_eur_leave23andme_20220403_1e-5.tsv
A2_ALL_his_leave_23andme
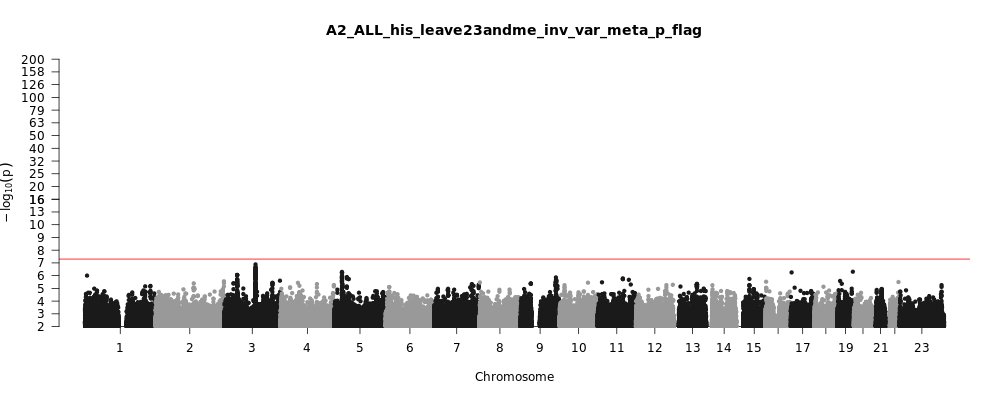
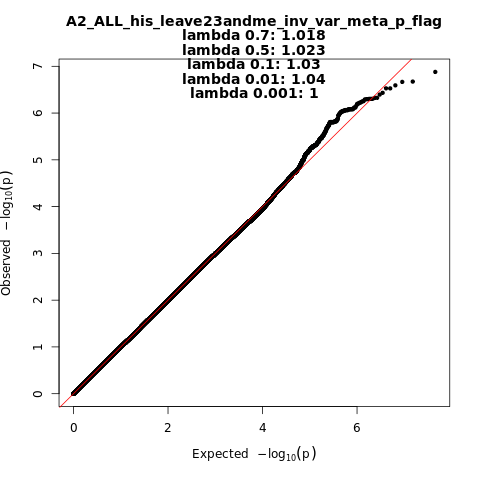
Phenotype
Very severe respiratory confirmed covid vs. population
Population
his
Total Cases
769
Total Controls
6209
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BRACOVID_AMR | 539 | 1149 |
| MexGen-COVID_AMR | 230 | 5060 |
Downloads
GRCh37 liftover: COVID19_HGI_A2_ALL_his_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_A2_ALL_his_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_A2_ALL_his_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_A2_ALL_his_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_A2_ALL_his_leave23andme_20220403_1e-5.tsv
A2_ALL_sas_leave_23andme
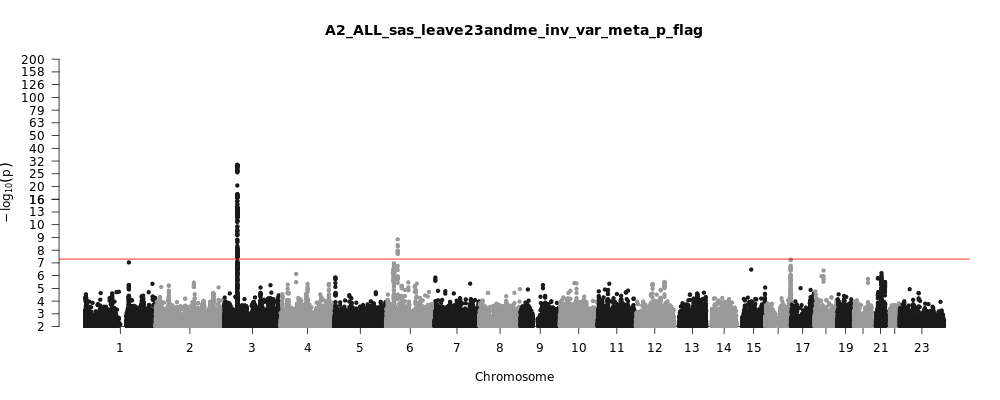
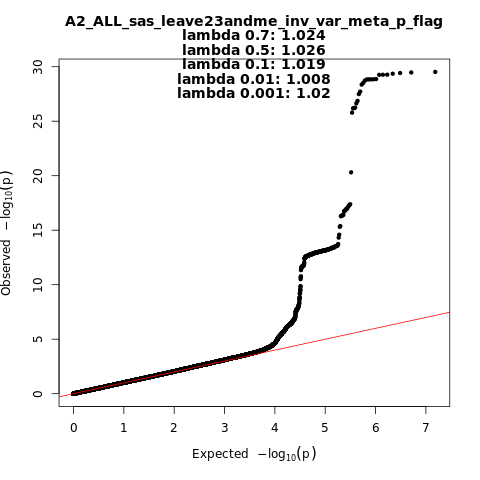
Phenotype
Very severe respiratory confirmed covid vs. population
Population
sas
Total Cases
980
Total Controls
47696
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| GNH_SAS | 192 | 43998 |
| genomicc_SAS | 788 | 3698 |
Downloads
GRCh37 liftover: COVID19_HGI_A2_ALL_sas_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_A2_ALL_sas_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_A2_ALL_sas_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_A2_ALL_sas_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_A2_ALL_sas_leave23andme_20220403_1e-5.tsv
B1_ALL_leave_23andme
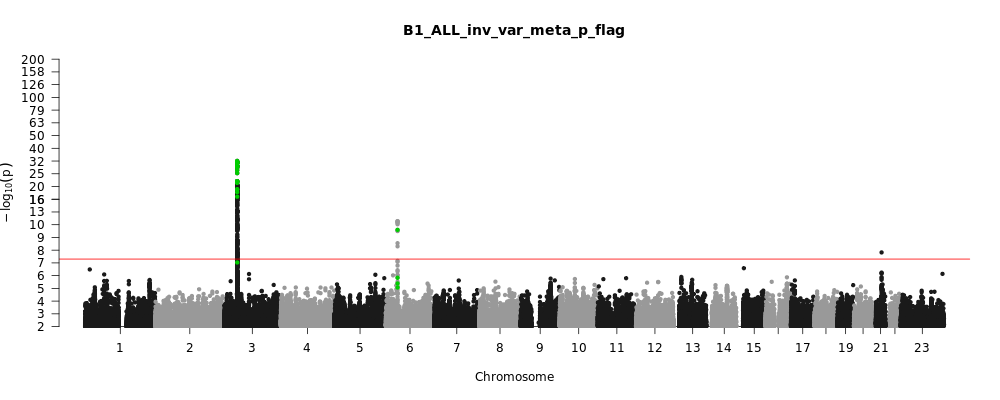
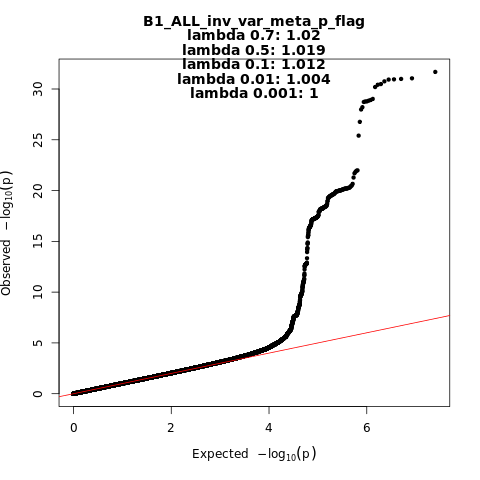
Phenotype
Hospitalized covid vs. not hospitalized covid
Population
All
Total Cases
16512
Total Controls
71321
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| CHOP_CAG_AFR | 147 | 412 |
| FHoGID_EUR | 470 | 280 |
| IranCovid_MID | 210 | 183 |
| BQC19_EUR | 654 | 406 |
| BelCovid_EUR | 504 | 299 |
| BioVU_EUR | 285 | 1324 |
| BoSCO_EUR | 425 | 743 |
| CCPM_EUR | 189 | 1242 |
| CGEN_EUR | 267 | 307 |
| ChiCovid_AMR | 143 | 420 |
| FinnGen_FIN | 437 | 3105 |
| GNH_SAS | 423 | 43767 |
| IND_GJ_COVID19_SAS | 411 | 147 |
| INMUNGEN_CoV2_EUR | 347 | 65 |
| JSA-COVID19_EAS | 461 | 1193 |
| MexCovid_AMR | 323 | 198 |
| PHBB_EUR | 318 | 732 |
| QGP_MID | 191 | 1865 |
| SCOURGE_EUR | 5934 | 3355 |
| UCLA_AMR | 166 | 405 |
| UCLA_EUR | 150 | 696 |
| UKBB_AFR | 137 | 286 |
| UKBB_CSA | 172 | 482 |
| UKBB_EUR | 3748 | 9409 |
Downloads
GRCh37 liftover: COVID19_HGI_B1_ALL_leave_23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_B1_ALL_leave_23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_B1_ALL_leave_23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_B1_ALL_leave_23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_B1_ALL_leave_23andme_20220403_1e-5.tsv
B2_ALL_leave_23andme
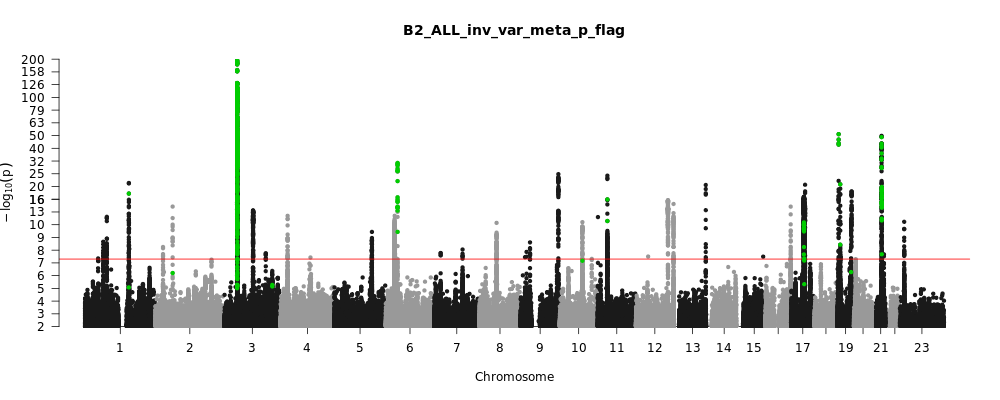
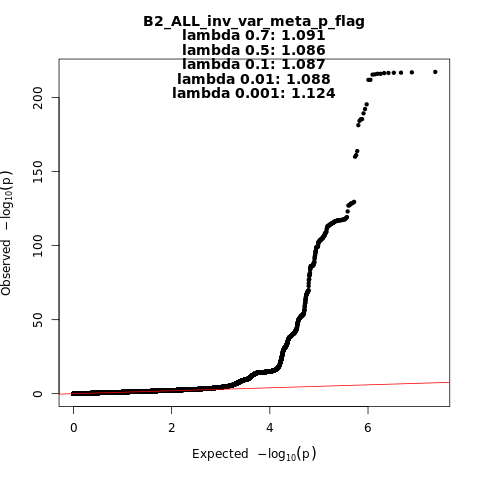
Phenotype
Hospitalized covid vs. population
Population
All
Total Cases
44986
Total Controls
2356386
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ANCESTRY_Freeze_Four_AFR | 135 | 5688 |
| ANCESTRY_Freeze_Four_AMR | 199 | 12323 |
| ANCESTRY_Freeze_Four_EUR | 1484 | 113882 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| BRACOVID_AMR | 853 | 835 |
| CU_AFR | 304 | 2610 |
| CU_EUR | 453 | 2149 |
| Corea_EAS | 69 | 6500 |
| DECODE_EUR | 89 | 274322 |
| EstBB_EUR | 352 | 196455 |
| GENCOVID_EUR | 1287 | 2443 |
| GHS_Freeze_145_EUR | 773 | 108168 |
| Generation_Scotland_EUR | 554 | 18683 |
| LGDB_EUR | 57 | 1531 |
| MVP_AFR | 1300 | 98129 |
| MVP_AMR | 517 | 43062 |
| MVP_EUR | 2417 | 366449 |
| PMBB_AFR | 126 | 8551 |
| SPGRX_EUR | 311 | 302 |
| idipaz24genetics_EUR | 106 | 75 |
| ArmCovid_OTH | 175 | 160 |
| BQC19_EUR | 654 | 914 |
| BelCovid_EUR | 505 | 1477 |
| BioVU_EUR | 285 | 68064 |
| BoSCO_EUR | 425 | 984 |
| CCPM_EUR | 189 | 25696 |
| COMRI_EUR | 120 | 333 |
| Charite_EUR | 198 | 865 |
| ChiCovid_AMR | 143 | 522 |
| FinnGen_FIN | 437 | 337685 |
| FrenchCovid_EUR | 456 | 982 |
| Genetics_COVID19_Korea_EAS | 932 | 19672 |
| HOSTAGE_EUR | 3117 | 12483 |
| INMUNGEN_CoV2_EUR | 347 | 723 |
| Ioannina_EUR | 321 | 852 |
| JapanTaskForce_EAS | 998 | 3317 |
| JorCovid_MID | 523 | 529 |
| KazCovid_EAS | 148 | 166 |
| MexGen-COVID_AMR | 876 | 5060 |
| PHBB_EUR | 318 | 31558 |
| POLISH_COVID_WGS_EUR | 359 | 871 |
| PakCovid_CSA | 180 | 174 |
| QGP_MID | 191 | 13868 |
| SCOURGE_EUR | 5934 | 8810 |
| UCLA_AMR | 166 | 4686 |
| UCLA_EUR | 150 | 21234 |
| UKBB_AFR | 137 | 6499 |
| UKBB_CSA | 172 | 8704 |
| UKBB_EUR | 3748 | 416783 |
| CHOP_CAG_AFR | 147 | 412 |
| FHoGID_EUR | 470 | 280 |
| IranCovid_MID | 210 | 183 |
| CGEN_EUR | 267 | 307 |
| GNH_SAS | 423 | 43767 |
| IND_GJ_COVID19_SAS | 411 | 147 |
| JSA-COVID19_EAS | 461 | 1193 |
| MexCovid_AMR | 323 | 198 |
| SaudiHumanGenomeProgram_ARAB | 147 | 768 |
| Vanda_EUR | 58 | 900 |
| Egypt_hgCOVID_hub_MID | 699 | 472 |
| SweCovid_EUR | 181 | 3748 |
| genomicc_AFR | 440 | 1336 |
| genomicc_EAS | 274 | 352 |
| genomicc_EUR | 5989 | 41384 |
| genomicc_SAS | 788 | 3698 |
Downloads
GRCh37 liftover: COVID19_HGI_B2_ALL_leave_23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_B2_ALL_leave_23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_B2_ALL_leave_23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_B2_ALL_leave_23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_B2_ALL_leave_23andme_20220403_1e-5.tsv
B2_ALL_afr_leave_23andme
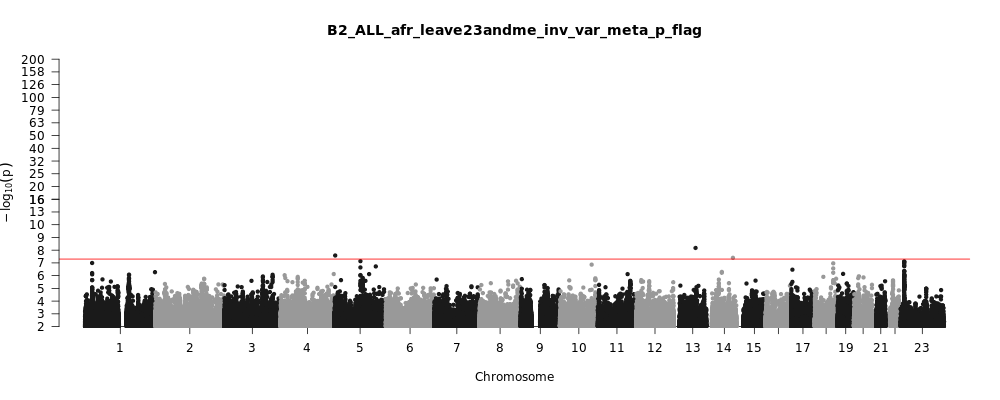
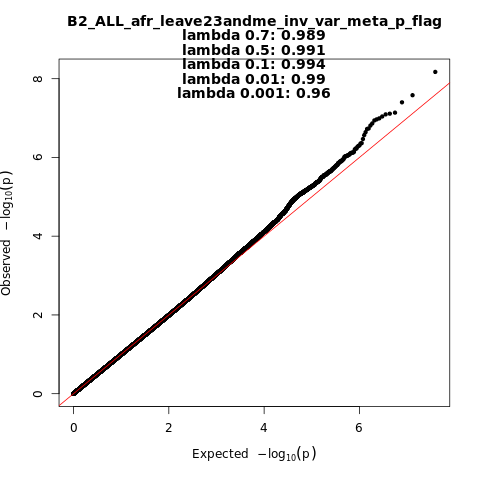
Phenotype
Hospitalized covid vs. population
Population
afr
Total Cases
2589
Total Controls
123225
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ANCESTRY_Freeze_Four_AFR | 135 | 5688 |
| CU_AFR | 304 | 2610 |
| MVP_AFR | 1300 | 98129 |
| PMBB_AFR | 126 | 8551 |
| UKBB_AFR | 137 | 6499 |
| CHOP_CAG_AFR | 147 | 412 |
| genomicc_AFR | 440 | 1336 |
Downloads
GRCh37 liftover: COVID19_HGI_B2_ALL_afr_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_B2_ALL_afr_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_B2_ALL_afr_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_B2_ALL_afr_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_B2_ALL_afr_leave23andme_20220403_1e-5.tsv
B2_ALL_eas_leave_23andme
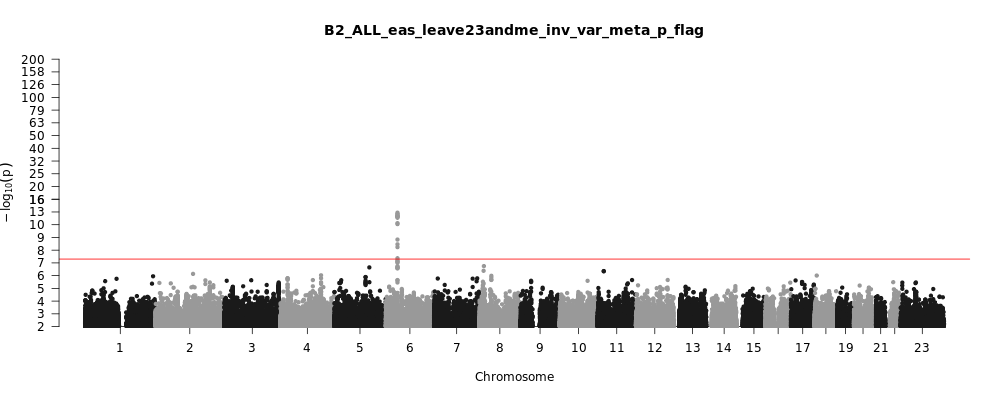
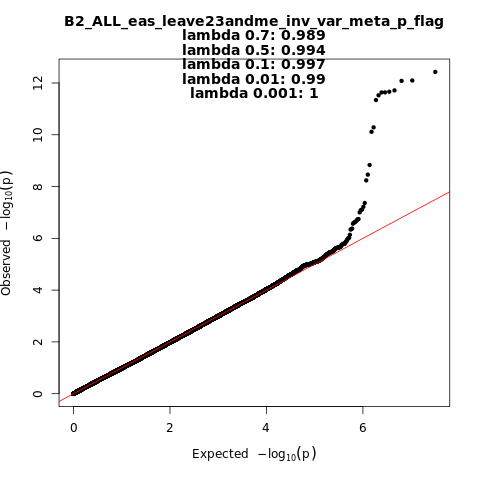
Phenotype
Hospitalized covid vs. population
Population
eas
Total Cases
2882
Total Controls
31200
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| Corea_EAS | 69 | 6500 |
| Genetics_COVID19_Korea_EAS | 932 | 19672 |
| JapanTaskForce_EAS | 998 | 3317 |
| KazCovid_EAS | 148 | 166 |
| JSA-COVID19_EAS | 461 | 1193 |
| genomicc_EAS | 274 | 352 |
Downloads
GRCh37 liftover: COVID19_HGI_B2_ALL_eas_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_B2_ALL_eas_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_B2_ALL_eas_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_B2_ALL_eas_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_B2_ALL_eas_leave23andme_20220403_1e-5.tsv
B2_ALL_eur_leave_23andme
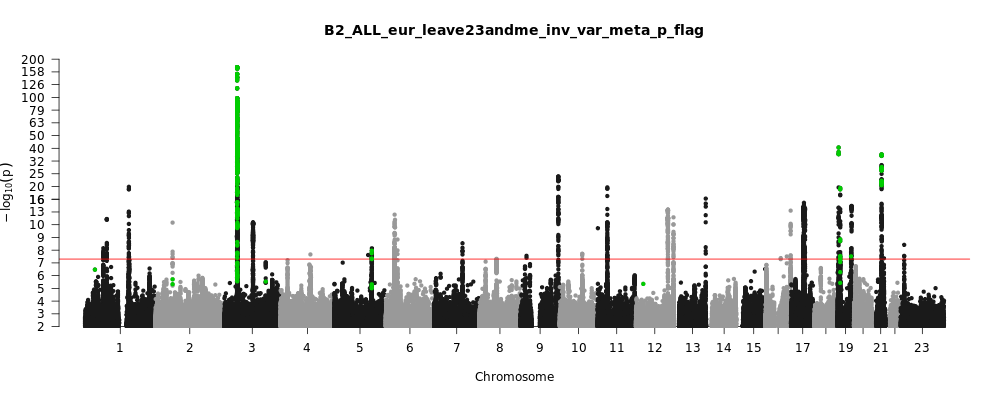
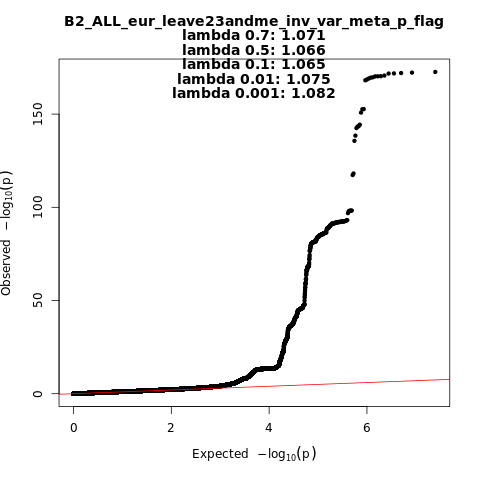
Phenotype
Hospitalized covid vs. population
Population
eur
Total Cases
32519
Total Controls
2062805
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ANCESTRY_Freeze_Four_EUR | 1484 | 113882 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| CU_EUR | 453 | 2149 |
| DECODE_EUR | 89 | 274322 |
| EstBB_EUR | 352 | 196455 |
| GENCOVID_EUR | 1287 | 2443 |
| GHS_Freeze_145_EUR | 773 | 108168 |
| Generation_Scotland_EUR | 554 | 18683 |
| LGDB_EUR | 57 | 1531 |
| MVP_EUR | 2417 | 366449 |
| SPGRX_EUR | 311 | 302 |
| idipaz24genetics_EUR | 106 | 75 |
| BQC19_EUR | 654 | 914 |
| BelCovid_EUR | 505 | 1477 |
| BioVU_EUR | 285 | 68064 |
| BoSCO_EUR | 425 | 984 |
| CCPM_EUR | 189 | 25696 |
| COMRI_EUR | 120 | 333 |
| Charite_EUR | 198 | 865 |
| FinnGen_FIN | 437 | 337685 |
| FrenchCovid_EUR | 456 | 982 |
| HOSTAGE_EUR | 3117 | 12483 |
| INMUNGEN_CoV2_EUR | 347 | 723 |
| Ioannina_EUR | 321 | 852 |
| PHBB_EUR | 318 | 31558 |
| POLISH_COVID_WGS_EUR | 359 | 871 |
| SCOURGE_EUR | 5934 | 8810 |
| UCLA_EUR | 150 | 21234 |
| UKBB_EUR | 3748 | 416783 |
| FHoGID_EUR | 470 | 280 |
| CGEN_EUR | 267 | 307 |
| Vanda_EUR | 58 | 900 |
| SweCovid_EUR | 181 | 3748 |
| genomicc_EUR | 5989 | 41384 |
Downloads
GRCh37 liftover: COVID19_HGI_B2_ALL_eur_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_B2_ALL_eur_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_B2_ALL_eur_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_B2_ALL_eur_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_B2_ALL_eur_leave23andme_20220403_1e-5.tsv
B2_ALL_his_leave_23andme
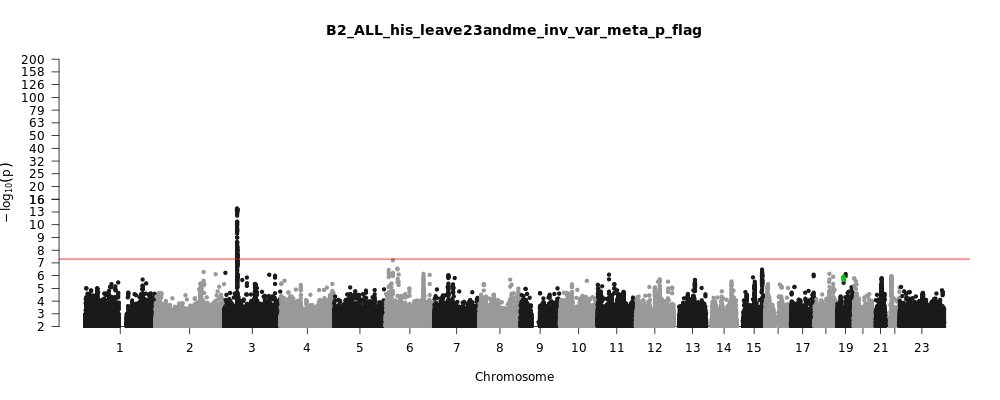
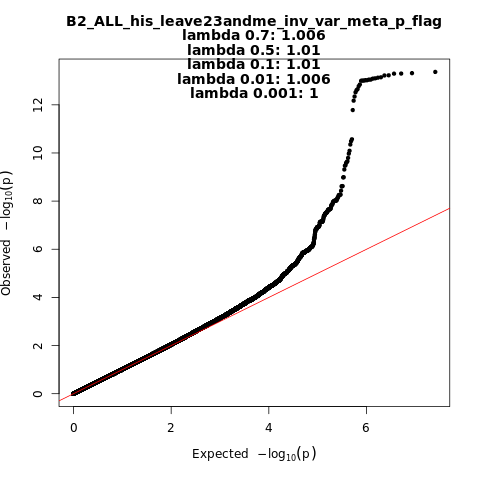
Phenotype
Hospitalized covid vs. population
Population
his
Total Cases
3077
Total Controls
66686
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ANCESTRY_Freeze_Four_AMR | 199 | 12323 |
| BRACOVID_AMR | 853 | 835 |
| MVP_AMR | 517 | 43062 |
| ChiCovid_AMR | 143 | 522 |
| MexGen-COVID_AMR | 876 | 5060 |
| UCLA_AMR | 166 | 4686 |
| MexCovid_AMR | 323 | 198 |
Downloads
GRCh37 liftover: COVID19_HGI_B2_ALL_his_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_B2_ALL_his_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_B2_ALL_his_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_B2_ALL_his_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_B2_ALL_his_leave23andme_20220403_1e-5.tsv
B2_ALL_sas_leave_23andme
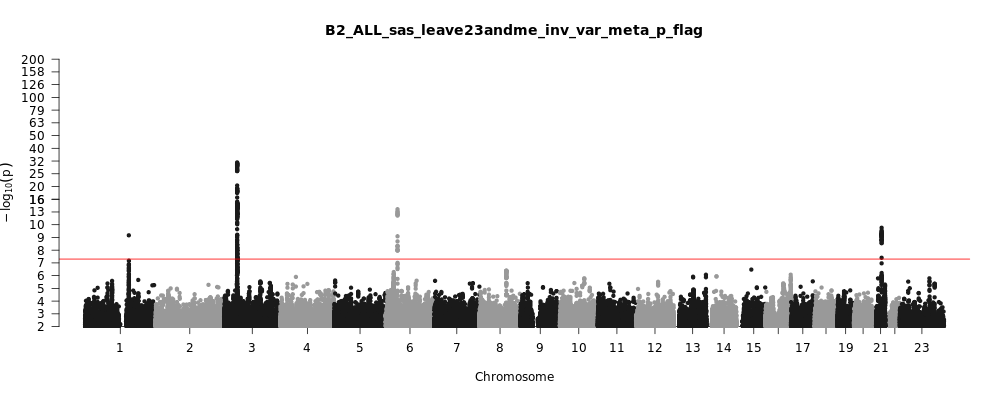
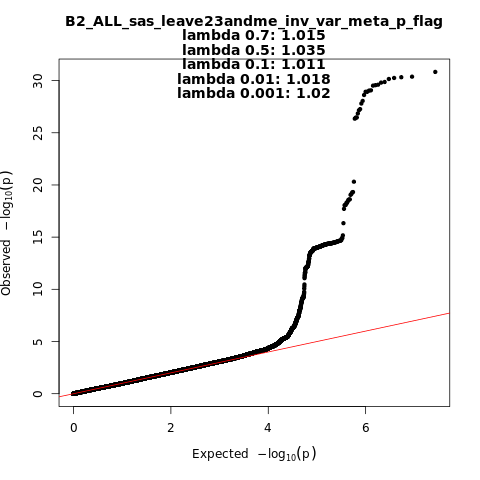
Phenotype
Hospitalized covid vs. population
Population
sas
Total Cases
1622
Total Controls
47612
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| GNH_SAS | 423 | 43767 |
| IND_GJ_COVID19_SAS | 411 | 147 |
| genomicc_SAS | 788 | 3698 |
Downloads
GRCh37 liftover: COVID19_HGI_B2_ALL_sas_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_B2_ALL_sas_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_B2_ALL_sas_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_B2_ALL_sas_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_B2_ALL_sas_leave23andme_20220403_1e-5.tsv
C2_ALL_leave_23andme
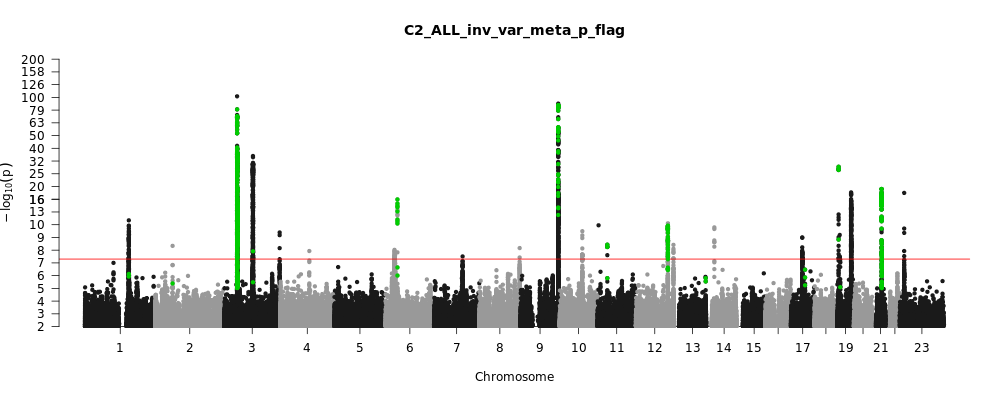
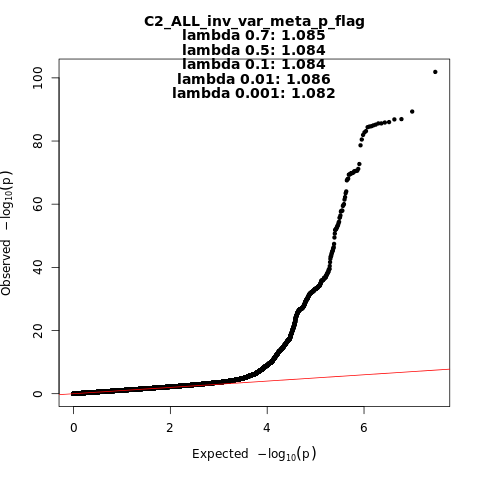
Phenotype
Covid vs. population
Population
All
Total Cases
159840
Total Controls
2782977
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ACCOuNT_AFR | 57 | 799 |
| ANCESTRY_Freeze_Four_AFR | 1629 | 5688 |
| ANCESTRY_Freeze_Four_AMR | 3752 | 12323 |
| ANCESTRY_Freeze_Four_EAS | 120 | 570 |
| ANCESTRY_Freeze_Four_EUR | 25353 | 113882 |
| CU_AFR | 332 | 2610 |
| CU_EUR | 508 | 2149 |
| Corea_EAS | 108 | 6500 |
| Coronagenes_EUR | 181 | 734 |
| EXCEED_EUR | 236 | 1083 |
| EstBB_EUR | 4070 | 189406 |
| GENCOVID_EUR | 1697 | 2443 |
| GFG_EUR | 136 | 4978 |
| GHS_Freeze_145_AFR | 128 | 3050 |
| GHS_Freeze_145_AMR | 88 | 1307 |
| GHS_Freeze_145_EUR | 5276 | 108168 |
| Generation_Scotland_EUR | 1712 | 17525 |
| HUNT_EUR | 283 | 52072 |
| LGDB_EUR | 275 | 1313 |
| Lifelines_EUR | 895 | 26600 |
| MGI_EUR | 122 | 51458 |
| MGI_AFR | 111 | 2989 |
| MOBA_EUR | 391 | 56038 |
| MVP_AFR | 4893 | 94556 |
| MVP_AMR | 2497 | 41100 |
| MVP_EUR | 11778 | 357198 |
| NTR_EUR | 228 | 5265 |
| PMBB_AFR | 379 | 8298 |
| PMBB_EUR | 60 | 9702 |
| SPGRX_EUR | 362 | 302 |
| Stanford_EUR | 169 | 190 |
| TOPMed_CHRIS10K_EUR | 92 | 2373 |
| TOPMed_Gardena_EUR | 452 | 458 |
| WGHS_EUR | 225 | 11887 |
| genomicsengland100kgp_EUR | 822 | 44369 |
| genomicsengland100kgp_SAS | 235 | 4205 |
| thaicovid_EAS | 191 | 113 |
| ABCD_EUR | 234 | 3135 |
| ALSPAC_G1_EUR | 523 | 4199 |
| BQC19_EUR | 1060 | 508 |
| BelCovid_EUR | 804 | 1477 |
| BioVU_EUR | 1592 | 68064 |
| BoSCO_EUR | 1168 | 984 |
| CCHC_HIS | 196 | 3039 |
| CCPM_AMR | 320 | 3074 |
| CCPM_EUR | 1496 | 24389 |
| ChiCovid_AMR | 705 | 522 |
| DBDS_EUR | 5554 | 87127 |
| ERACORE_EUR | 210 | 1547 |
| Egypt_hgCOVID_hub_MID | 710 | 472 |
| FinnGen_FIN | 4814 | 337685 |
| GCAT_EUR | 710 | 4278 |
| GNH_SAS | 7922 | 36268 |
| Genotek_EUR | 6959 | 18720 |
| HOSTAGE_EUR | 3097 | 10327 |
| Helix_EUR | 1055 | 10960 |
| INMUNGEN_CoV2_EUR | 412 | 723 |
| JapanTaskForce_EAS | 2433 | 3317 |
| JorCovid_MID | 529 | 529 |
| MexGen-COVID_AMR | 939 | 5060 |
| PHBB_AFR | 160 | 1909 |
| PHBB_AMR | 270 | 1970 |
| PHBB_EUR | 1059 | 31558 |
| POLISH_COVID_WGS_EUR | 235 | 998 |
| QGP_MID | 2056 | 12003 |
| SCOURGE_EUR | 9301 | 5455 |
| SINAI_COVID_AFR | 74 | 143 |
| SINAI_COVID_AMR | 128 | 123 |
| SINAI_COVID_EUR | 1329 | 8940 |
| TIKOCO_EUR | 572 | 3436 |
| UCLA_AFR | 152 | 1585 |
| UCLA_AMR | 571 | 4281 |
| UCLA_EAS | 140 | 2835 |
| UCLA_EUR | 846 | 20538 |
| UKBB_AFR | 459 | 6177 |
| UKBB_AMR | 58 | 922 |
| UKBB_CSA | 735 | 8141 |
| UKBB_EAS | 113 | 2596 |
| UKBB_EUR | 14539 | 405992 |
| UKBB_MID | 97 | 1502 |
| INTERVAL_EUR | 2098 | 39733 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| BRACOVID_AMR | 853 | 835 |
| DECODE_EUR | 89 | 274322 |
| idipaz24genetics_EUR | 106 | 75 |
| ArmCovid_OTH | 175 | 160 |
| COMRI_EUR | 120 | 333 |
| Charite_EUR | 198 | 865 |
| FrenchCovid_EUR | 456 | 982 |
| Genetics_COVID19_Korea_EAS | 932 | 19672 |
| Ioannina_EUR | 321 | 852 |
| KazCovid_EAS | 148 | 166 |
| PakCovid_CSA | 180 | 174 |
| SaudiHumanGenomeProgram_ARAB | 147 | 768 |
| Vanda_EUR | 58 | 900 |
| SweCovid_EUR | 181 | 3748 |
| genomicc_AFR | 440 | 1336 |
| genomicc_EAS | 274 | 352 |
| genomicc_EUR | 5989 | 41384 |
| genomicc_SAS | 788 | 3698 |
Downloads
GRCh37 liftover: COVID19_HGI_C2_ALL_leave_23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_C2_ALL_leave_23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_C2_ALL_leave_23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_C2_ALL_leave_23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_C2_ALL_leave_23andme_20220403_1e-5.tsv
C2_ALL_afr_leave_23andme
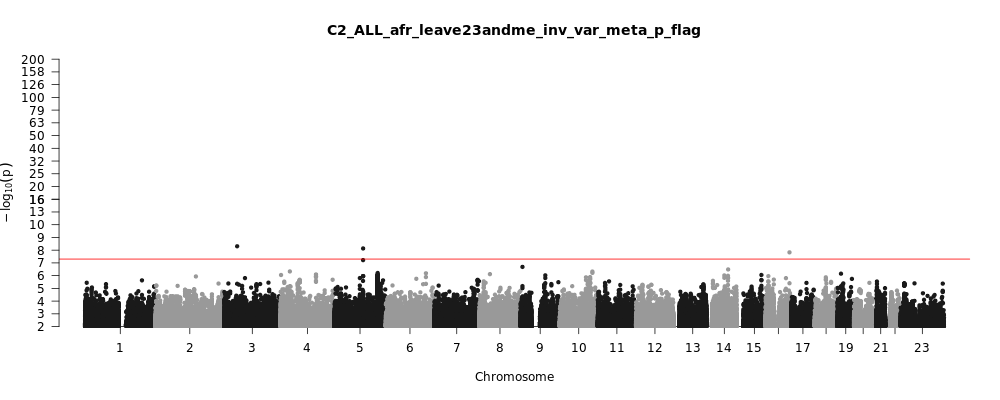
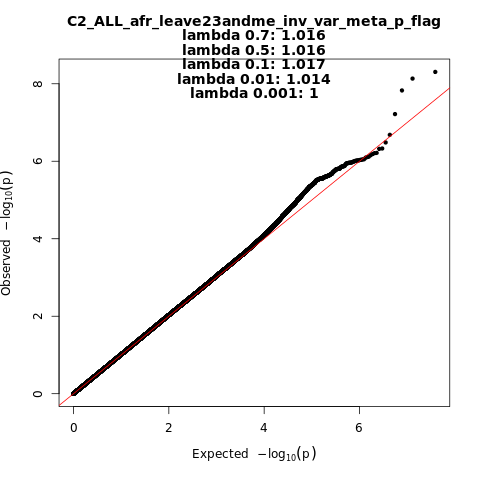
Phenotype
Covid vs. population
Population
afr
Total Cases
8814
Total Controls
129140
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ACCOuNT_AFR | 57 | 799 |
| ANCESTRY_Freeze_Four_AFR | 1629 | 5688 |
| CU_AFR | 332 | 2610 |
| GHS_Freeze_145_AFR | 128 | 3050 |
| MGI_AFR | 111 | 2989 |
| MVP_AFR | 4893 | 94556 |
| PMBB_AFR | 379 | 8298 |
| PHBB_AFR | 160 | 1909 |
| SINAI_COVID_AFR | 74 | 143 |
| UCLA_AFR | 152 | 1585 |
| UKBB_AFR | 459 | 6177 |
| genomicc_AFR | 440 | 1336 |
Downloads
GRCh37 liftover: COVID19_HGI_C2_ALL_afr_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_C2_ALL_afr_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_C2_ALL_afr_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_C2_ALL_afr_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_C2_ALL_afr_leave23andme_20220403_1e-5.tsv
C2_ALL_eas_leave_23andme
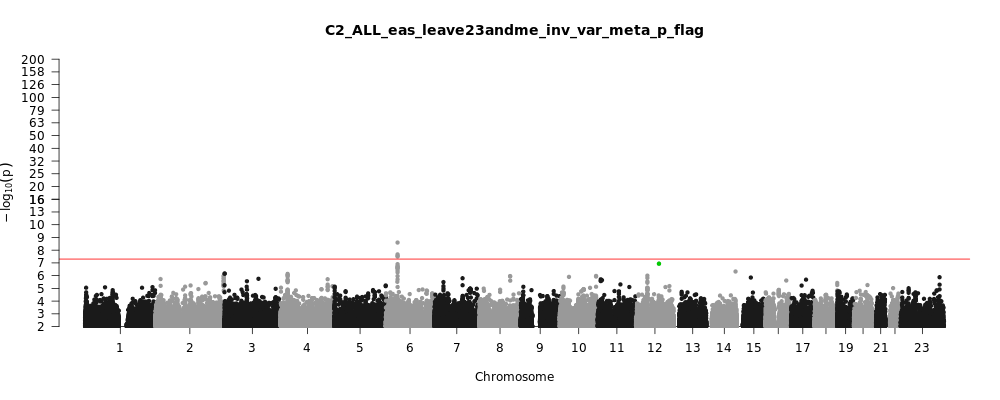
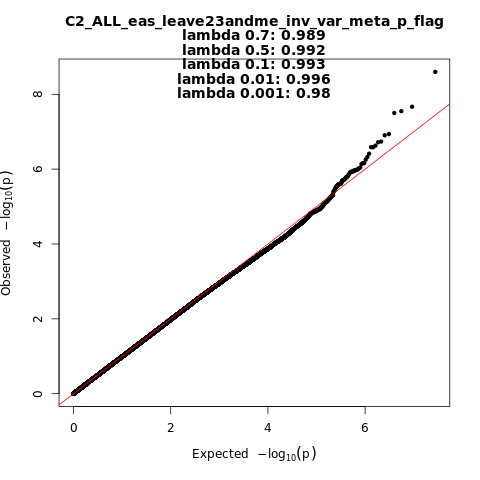
Phenotype
Covid vs. population
Population
eas
Total Cases
4459
Total Controls
36121
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ANCESTRY_Freeze_Four_EAS | 120 | 570 |
| Corea_EAS | 108 | 6500 |
| thaicovid_EAS | 191 | 113 |
| JapanTaskForce_EAS | 2433 | 3317 |
| UCLA_EAS | 140 | 2835 |
| UKBB_EAS | 113 | 2596 |
| Genetics_COVID19_Korea_EAS | 932 | 19672 |
| KazCovid_EAS | 148 | 166 |
| genomicc_EAS | 274 | 352 |
Downloads
GRCh37 liftover: COVID19_HGI_C2_ALL_eas_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_C2_ALL_eas_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_C2_ALL_eas_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_C2_ALL_eas_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_C2_ALL_eas_leave23andme_20220403_1e-5.tsv
C2_ALL_eur_leave_23andme
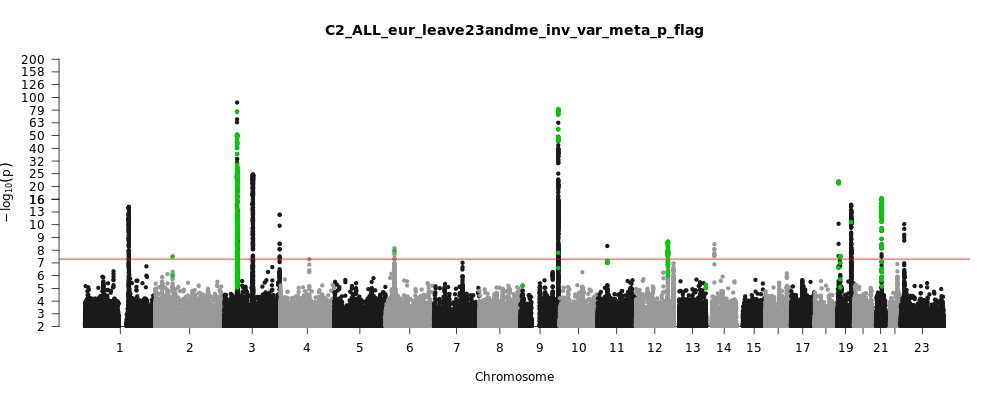
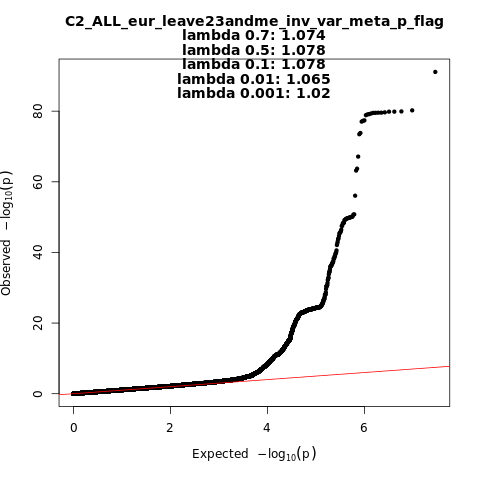
Phenotype
Covid vs. population
Population
eur
Total Cases
122616
Total Controls
2475240
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ANCESTRY_Freeze_Four_EUR | 25353 | 113882 |
| CU_EUR | 508 | 2149 |
| Coronagenes_EUR | 181 | 734 |
| EXCEED_EUR | 236 | 1083 |
| EstBB_EUR | 4070 | 189406 |
| GENCOVID_EUR | 1697 | 2443 |
| GFG_EUR | 136 | 4978 |
| GHS_Freeze_145_EUR | 5276 | 108168 |
| Generation_Scotland_EUR | 1712 | 17525 |
| HUNT_EUR | 283 | 52072 |
| LGDB_EUR | 275 | 1313 |
| Lifelines_EUR | 895 | 26600 |
| MGI_EUR | 122 | 51458 |
| MOBA_EUR | 391 | 56038 |
| MVP_EUR | 11778 | 357198 |
| NTR_EUR | 228 | 5265 |
| PMBB_EUR | 60 | 9702 |
| SPGRX_EUR | 362 | 302 |
| Stanford_EUR | 169 | 190 |
| TOPMed_CHRIS10K_EUR | 92 | 2373 |
| TOPMed_Gardena_EUR | 452 | 458 |
| WGHS_EUR | 225 | 11887 |
| genomicsengland100kgp_EUR | 822 | 44369 |
| ABCD_EUR | 234 | 3135 |
| ALSPAC_G1_EUR | 523 | 4199 |
| BQC19_EUR | 1060 | 508 |
| BelCovid_EUR | 804 | 1477 |
| BioVU_EUR | 1592 | 68064 |
| BoSCO_EUR | 1168 | 984 |
| CCPM_EUR | 1496 | 24389 |
| DBDS_EUR | 5554 | 87127 |
| ERACORE_EUR | 210 | 1547 |
| FinnGen_FIN | 4814 | 337685 |
| GCAT_EUR | 710 | 4278 |
| Genotek_EUR | 6959 | 18720 |
| HOSTAGE_EUR | 3097 | 10327 |
| Helix_EUR | 1055 | 10960 |
| INMUNGEN_CoV2_EUR | 412 | 723 |
| PHBB_EUR | 1059 | 31558 |
| POLISH_COVID_WGS_EUR | 235 | 998 |
| SCOURGE_EUR | 9301 | 5455 |
| SINAI_COVID_EUR | 1329 | 8940 |
| TIKOCO_EUR | 572 | 3436 |
| UCLA_EUR | 846 | 20538 |
| UKBB_EUR | 14539 | 405992 |
| INTERVAL_EUR | 2098 | 39733 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| DECODE_EUR | 89 | 274322 |
| idipaz24genetics_EUR | 106 | 75 |
| COMRI_EUR | 120 | 333 |
| Charite_EUR | 198 | 865 |
| FrenchCovid_EUR | 456 | 982 |
| Ioannina_EUR | 321 | 852 |
| Vanda_EUR | 58 | 900 |
| SweCovid_EUR | 181 | 3748 |
| genomicc_EUR | 5989 | 41384 |
Downloads
GRCh37 liftover: COVID19_HGI_C2_ALL_eur_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_C2_ALL_eur_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_C2_ALL_eur_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_C2_ALL_eur_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_C2_ALL_eur_leave23andme_20220403_1e-5.tsv
C2_ALL_his_leave_23andme
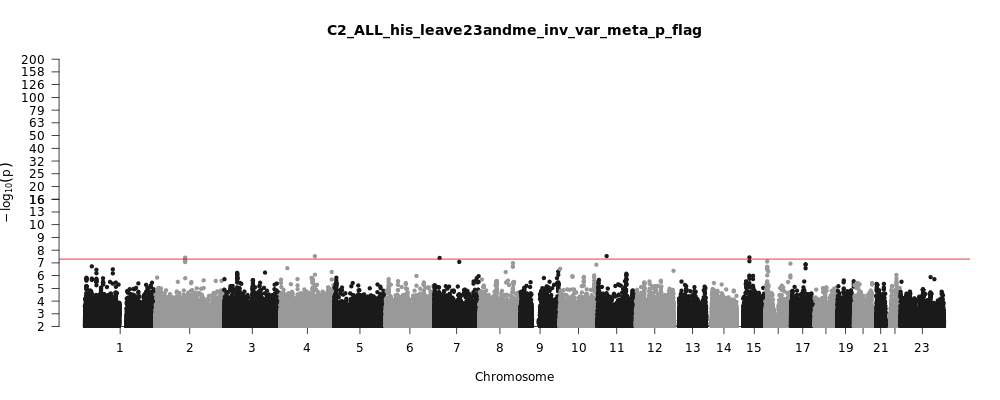
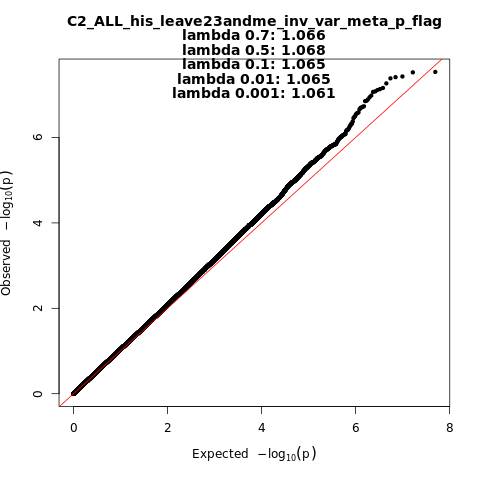
Phenotype
Covid vs. population
Population
his
Total Cases
10377
Total Controls
74556
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ANCESTRY_Freeze_Four_AMR | 3752 | 12323 |
| GHS_Freeze_145_AMR | 88 | 1307 |
| MVP_AMR | 2497 | 41100 |
| CCHC_HIS | 196 | 3039 |
| CCPM_AMR | 320 | 3074 |
| ChiCovid_AMR | 705 | 522 |
| MexGen-COVID_AMR | 939 | 5060 |
| PHBB_AMR | 270 | 1970 |
| SINAI_COVID_AMR | 128 | 123 |
| UCLA_AMR | 571 | 4281 |
| UKBB_AMR | 58 | 922 |
| BRACOVID_AMR | 853 | 835 |
Downloads
GRCh37 liftover: COVID19_HGI_C2_ALL_his_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_C2_ALL_his_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_C2_ALL_his_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_C2_ALL_his_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_C2_ALL_his_leave23andme_20220403_1e-5.tsv
C2_ALL_sas_leave_23andme
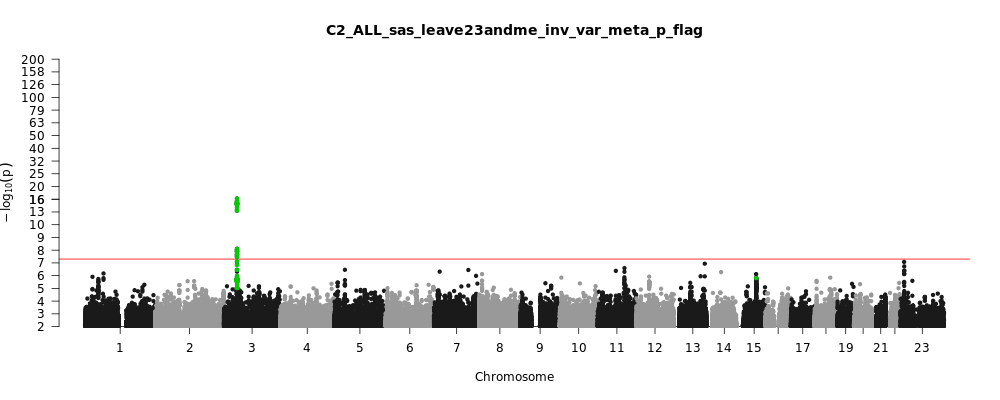
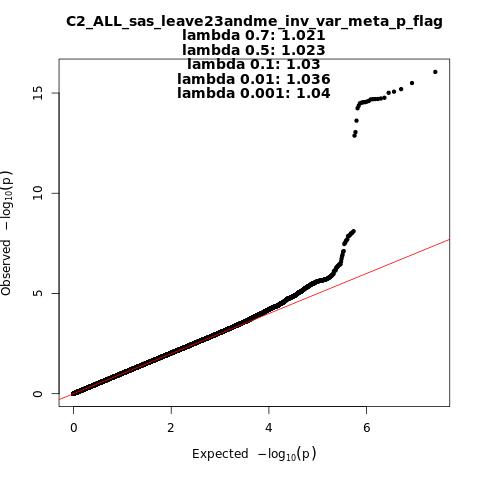
Phenotype
Covid vs. population
Population
sas
Total Cases
8945
Total Controls
44171
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| genomicsengland100kgp_SAS | 235 | 4205 |
| GNH_SAS | 7922 | 36268 |
| genomicc_SAS | 788 | 3698 |
Downloads
GRCh37 liftover: COVID19_HGI_C2_ALL_sas_leave23andme_20220403_GRCh37.tsv.gz
GRCh37 (.tbi): COVID19_HGI_C2_ALL_sas_leave23andme_20220403_GRCh37.tsv.gz.tbi
GRCh38: COVID19_HGI_C2_ALL_sas_leave23andme_20220403.tsv.gz
GRCh38 (.tbi): COVID19_HGI_C2_ALL_sas_leave23andme_20220403.tsv.gz.tbi
GRCh38 (filtered): COVID19_HGI_C2_ALL_sas_leave23andme_20220403_1e-5.tsv
